Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum)
- PMID: 24043829
- PMCID: PMC3791769
- DOI: 10.1073/pnas.1315760110
Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum)
Abstract
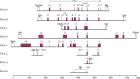
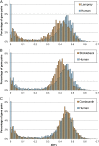
Cyclostomes, comprising jawless vertebrates such as lampreys and hagfishes, are the sister group of living jawed vertebrates (gnathostomes) and hence an important group for understanding the origin and diversity of vertebrates. In vertebrates and other metazoans, Hox genes determine cell fate along the anteroposterior axis of embryos and are implicated in driving morphological diversity. Invertebrates contain a single Hox cluster (either intact or fragmented), whereas elephant shark, coelacanth, and tetrapods contain four Hox clusters owing to two rounds of whole-genome duplication ("1R" and "2R") during early vertebrate evolution. By contrast, most teleost fishes contain up to eight Hox clusters because of an additional "teleost-specific" genome duplication event. By sequencing bacterial artificial chromosome (BAC) clones and the whole genome, here we provide evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). This suggests that the lamprey lineage has experienced an additional genome duplication after 1R and 2R. The relative age of lamprey and human paralogs supports this hypothesis. Compared with gnathostome Hox clusters, lamprey Hox clusters are unusually large. Several conserved noncoding elements (CNEs) were predicted in the Hox clusters of lamprey, elephant shark, and human. Transgenic zebrafish assay indicated the potential of CNEs to function as enhancers. Interestingly, CNEs in individual lamprey Hox clusters are frequently conserved in multiple Hox clusters in elephant shark and human, implying a many-to-many orthology relationship between lamprey and gnathostome Hox clusters. Such a relationship suggests that the first two rounds of genome duplication may have occurred independently in the lamprey and gnathostome lineages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Carroll SB, Weatherbee SD, Langeland JA. Homeotic genes and the regulation and evolution of insect wing number. Nature. 1995;375(6526):58–61. - PubMed
-
- Holland PW, Garcia-Fernàndez J. Hox genes and chordate evolution. Dev Biol. 1996;173(2):382–395. - PubMed
-
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134(14):2549–2560. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

