Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory
- PMID: 24043832
- PMCID: PMC3791776
- DOI: 10.1073/pnas.1309676110
Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory
Abstract
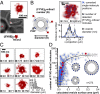
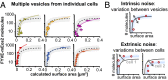
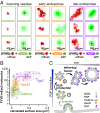
Cells tightly regulate trafficking of intracellular organelles, but a deeper understanding of this process is technically limited by our inability to track the molecular composition of individual organelles below the diffraction limit in size. Here we develop a technique for intracellularly calibrated superresolution microscopy that can measure the size of individual organelles as well as accurately count absolute numbers of molecules, by correcting for undercounting owing to immature fluorescent proteins and overcounting owing to fluorophore blinking. Using this technique, we characterized the size of individual vesicles in the yeast endocytic pathway and the number of accessible phosphatidylinositol 3-phosphate binding sites they contain. This analysis reveals a characteristic vesicle maturation trajectory of composition and size with both stochastic and regulated components. The trajectory displays some cell-to-cell variability, with smaller variation between organelles within the same cell. This approach also reveals mechanistic information on the order of events in this trajectory: Colocalization analysis with known markers of different vesicle maturation stages shows that phosphatidylinositol 3-phosphate production precedes fusion into larger endosomes. This single-organelle analysis can potentially be applied to a range of small organelles to shed light on their precise composition/structure relationships, the dynamics of their regulation, and the noise in these processes.
Keywords: GTPases; PALM; endocytosis; phosphoinositides; single-molecule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. - PubMed
-
- Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. - PubMed
-
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440(7082):358–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

