Structural features of Argonaute-GW182 protein interactions
- PMID: 24043833
- PMCID: PMC3791723
- DOI: 10.1073/pnas.1308510110
Structural features of Argonaute-GW182 protein interactions
Abstract
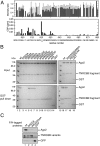
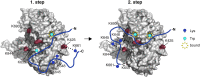
MicroRNAs (miRNAs) guide Argonaute (Ago) proteins to target mRNAs, leading to gene silencing. However, Ago proteins are not the actual mediators of gene silencing but interact with a member of the GW182 protein family (also known as GW proteins), which coordinates all downstream steps in gene silencing. GW proteins contain an N-terminal Ago-binding domain that is characterized by multiple GW repeats and a C-terminal silencing domain with several globular domains. Within the Ago-binding domain, Trp residues mediate the direct interaction with the Ago protein. Here, we have characterized the interaction of Ago proteins with GW proteins in molecular detail. Using biochemical and NMR experiments, we show that only a subset of Trp residues engage in Ago interactions. The Trp residues are located in intrinsically disordered regions, where flanking residues mediate additional weak interactions, that might explain the importance of specific tryptophans. Using cross-linking followed by mass spectrometry, we map the GW protein interactions with Ago2, which allows for structural modeling of Ago-GW182 interaction. Our data further indicate that the Ago-GW protein interaction might be a two-step process involving the sequential binding of two tryptophans separated by a spacer with a minimal length of 10 aa.
Keywords: RNA interference; RNAi; gene regulation; small RNA–mediated gene silencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. - PubMed
-
- Ender C, Meister G. Argonaute proteins at a glance. J Cell Sci. 2010;123(Pt 11):1819–1823. - PubMed
-
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457(7228):405–412. - PubMed
-
- Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

