Implementation of cell-free biological networks at steady state
- PMID: 24043836
- PMCID: PMC3791785
- DOI: 10.1073/pnas.1311166110
Implementation of cell-free biological networks at steady state
Abstract
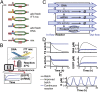
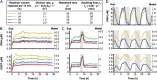
Living cells maintain a steady state of biochemical reaction rates by exchanging energy and matter with the environment. These exchanges usually do not occur in in vitro systems, which consequently go to chemical equilibrium. This in turn has severely constrained the complexity of biological networks that can be implemented in vitro. We developed nanoliter-scale microfluidic reactors that exchange reagents at dilution rates matching those of dividing bacteria. In these reactors we achieved transcription and translation at steady state for 30 h and implemented diverse regulatory mechanisms on the transcriptional, translational, and posttranslational levels, including RNA polymerases, transcriptional repression, translational activation, and proteolysis. We constructed and implemented an in vitro genetic oscillator and mapped its phase diagram showing that steady-state conditions were necessary to produce oscillations. This reactor-based approach will allow testing of whether fundamental limits exist to in vitro network complexity.
Keywords: cell-free protein synthesis; computational biology; minimal artificial cell; synthetic biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320(5877):789–792. - PubMed
-
- Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19(8):751–755. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

