Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity
- PMID: 24043837
- PMCID: PMC3791737
- DOI: 10.1073/pnas.1303111110
Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity
Abstract
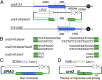
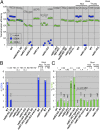
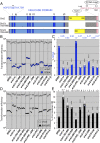
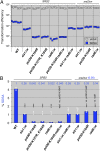
DNA damage alone or DNA replication fork arrest at damaged sites may induce DNA double-strand breaks and initiate homologous recombination. This event can result in a crossover with a homologous chromosome, causing loss of heterozygosity along the chromosome. It is known that Srs2 acts as an antirecombinase at the replication fork: it is recruited by the SUMO (a small ubiquitin-related modifier)-conjugated DNA-polymerase sliding clamp (PCNA) and interferes with Rad51/Rad52-mediated homologous recombination. Here, we report that Srs2 promotes another type of homologous recombination that produces noncrossover products only, in collaboration with PCNA and Rad51. Srs2 proteins lacking the Rad51-binding domain, PCNA-SUMO-binding motifs, or ATP hydrolysis-dependent DNA helicase activity reduce this noncrossover recombination. However, the removal of either the Rad51-binding domain or the PCNA-binding motif strongly increases crossovers. Srs2 gene mutations are epistatic to mutations in the PCNA modification-related genes encoding PCNA, Siz1 (a SUMO ligase) and Rad6 (a ubiquitin-conjugating protein). Knocking out RAD51 blocked this recombination but enhanced nonhomologous end-joining. We hypothesize that, during DNA double-strand break repair, Srs2 mediates collaboration between the Rad51 nucleofilament and PCNA-SUMO and directs the heteroduplex intermediate to DNA synthesis in a moving bubble. This Rad51/Rad52/Srs2/PCNA-mediated noncrossover pathway avoids both interchromosomal crossover and imprecise end-joining, two potential paths leading to loss of heterozygosity, and contributes to genome maintenance and human health.
Keywords: NHEJ; SDSA; bubble migration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair.FEMS Yeast Res. 2017 Mar 1;17(2):fow111. doi: 10.1093/femsyr/fow111. FEMS Yeast Res. 2017. PMID: 28011904 Free PMC article. Review.
-
DNA Damage Tolerance Pathway Choice Through Uls1 Modulation of Srs2 SUMOylation in Saccharomyces cerevisiae.Genetics. 2017 May;206(1):513-525. doi: 10.1534/genetics.116.196568. Epub 2017 Mar 24. Genetics. 2017. PMID: 28341648 Free PMC article.
-
Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases.Genetics. 2012 May;191(1):65-78. doi: 10.1534/genetics.112.139105. Epub 2012 Feb 23. Genetics. 2012. PMID: 22367032 Free PMC article.
-
The Main Role of Srs2 in DNA Repair Depends on Its Helicase Activity, Rather than on Its Interactions with PCNA or Rad51.mBio. 2018 Jul 17;9(4):e01192-18. doi: 10.1128/mBio.01192-18. mBio. 2018. PMID: 30018112 Free PMC article.
-
Multifaceted role of the Saccharomyces cerevisiae Srs2 helicase in homologous recombination regulation.Biochem Soc Trans. 2005 Dec;33(Pt 6):1447-50. doi: 10.1042/BST0331447. Biochem Soc Trans. 2005. PMID: 16246143 Review.
Cited by
-
Nonfilament-forming RecA dimer catalyzes homologous joint formation.Nucleic Acids Res. 2018 Nov 16;46(20):10855-10869. doi: 10.1093/nar/gky877. Nucleic Acids Res. 2018. PMID: 30285153 Free PMC article.
-
SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):12793-12798. doi: 10.1073/pnas.1716349115. Epub 2018 Nov 28. Proc Natl Acad Sci U S A. 2018. PMID: 30487218 Free PMC article.
-
Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair.FEMS Yeast Res. 2017 Mar 1;17(2):fow111. doi: 10.1093/femsyr/fow111. FEMS Yeast Res. 2017. PMID: 28011904 Free PMC article. Review.
-
DNA Damage Tolerance Pathway Choice Through Uls1 Modulation of Srs2 SUMOylation in Saccharomyces cerevisiae.Genetics. 2017 May;206(1):513-525. doi: 10.1534/genetics.116.196568. Epub 2017 Mar 24. Genetics. 2017. PMID: 28341648 Free PMC article.
-
Putting together and taking apart: assembly and disassembly of the Rad51 nucleoprotein filament in DNA repair and genome stability.Cell Stress. 2018 Mar 28;2(5):96-112. doi: 10.15698/cst2018.05.134. Cell Stress. 2018. PMID: 31225474 Free PMC article. Review.
References
-
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436(7049):428–433. - PubMed
-
- Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423(6937):305–309. - PubMed
-
- Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423(6937):309–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

