Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea
- PMID: 24043856
- PMCID: PMC3830477
- DOI: 10.1093/jxb/ert280
Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea
Abstract
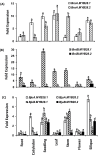
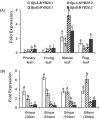
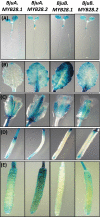
Glucosinolates are Capparales-specific secondary metabolites that have immense potential in human health and agriculture. Unlike Arabidopsis thaliana, our knowledge about glucosinolate regulators in the Brassica crops is sparse. In the current study, four MYB28 homologues were identified (BjuMYB28-1,-2,-3,-4) from the polyploid Brassica juncea, and the effects of allopolyploidization on the divergence of gene sequence, structure, function, and expression were assessed. The deduced protein sequences of the four BjuMYB28 genes showed 76.1-83.1% identity with the Arabidopsis MYB28. Phylogenetic analysis revealed that the four BjuMYB28 proteins have evolved via the hybridization and duplication processes forming the B. juncea genome (AABB) from B. rapa (AA) and B. nigra (BB), while retaining high levels of sequence conservation. Mutant complementation and over-expression studies in A. thaliana showed that all four BjuMYB28 genes encode functional MYB28 proteins and resulted in similar aliphatic glucosinolate composition and content. Detailed expression analysis using qRT-PCR assays and promoter-GUS lines revealed that the BjuMYB28 genes have both tissue- and cell-specific expression partitioning in B. juncea. The two B-genome origin BjuMYB28 genes had more abundant transcripts during the early stages of plant development than the A-genome origin genes. However, with the onset of the reproductive phase, expression levels of all four BjuMYB28 increased significantly, which may be necessary for producing and maintaining high amounts of aliphatic glucosinolates during the later stages of plant development. Taken together, our results suggest that the four MYB28 genes are differentially expressed and regulated in B. juncea to play discrete though overlapping roles in controlling aliphatic glucosinolate biosynthesis.
Keywords: Brassica juncea; MYB28; expression partitioning; glucosinolates; transcription factor..
Figures






References
-
- Adams KL. 2007. Evolution of duplicate gene expression in polyploid and hybrid plants. Journal of Heredity 98, 136–141 - PubMed
-
- Adams KL, Wendel JF. 2005. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology 8, 135–141 - PubMed
-
- Augustine R, Mukhopadhyay A, Bisht NC. 2013. Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea . Plant Biotechnology Journal 11, 855–866 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

