IL-33 markedly activates murine eosinophils by an NF-κB-dependent mechanism differentially dependent upon an IL-4-driven autoinflammatory loop
- PMID: 24043894
- PMCID: PMC3807853
- DOI: 10.4049/jimmunol.1301465
IL-33 markedly activates murine eosinophils by an NF-κB-dependent mechanism differentially dependent upon an IL-4-driven autoinflammatory loop
Abstract
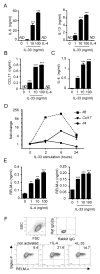
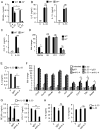
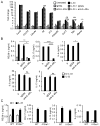
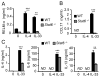
Eosinophils are major effector cells in type 2 inflammatory responses and become activated in response to IL-4 and IL-33, yet the molecular mechanisms and cooperative interaction between these cytokines remain unclear. Our objective was to investigate the molecular mechanism and cooperation of IL-4 and IL-33 in eosinophil activation. Eosinophils derived from bone marrow or isolated from Il5-transgenic mice were activated in the presence of IL-4 or IL-33 for 1 or 4 h, and the transcriptome was analyzed by RNA sequencing. The candidate genes were validated by quantitative PCR and ELISA. We demonstrated that murine-cultured eosinophils respond to IL-4 and IL-33 by phosphorylation of STAT-6 and NF-κB, respectively. RNA sequence analysis of murine-cultured eosinophils indicated that IL-33 induced 519 genes, whereas IL-4 induced only 28 genes, including 19 IL-33-regulated genes. Interestingly, IL-33 induced eosinophil activation via two distinct mechanisms, IL-4 independent and IL-4 secretion/autostimulation dependent. Anti-IL-4 or anti-IL-4Rα Ab-treated cultured and mature eosinophils, as well as Il4- or Stat6-deficient cultured eosinophils, had attenuated protein secretion of a subset of IL-33-induced genes, including Retnla and Ccl17. Additionally, IL-33 induced the rapid release of preformed IL-4 protein from eosinophils by a NF-κB-dependent mechanism. However, the induction of most IL-33-regulated transcripts (e.g., Il6 and Il13) was IL-4 independent and blocked by NF-κB inhibition. In conclusion, we have identified a novel activation pathway in murine eosinophils that is induced by IL-33 and differentially dependent upon an IL-4 auto-amplification loop.
Figures



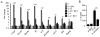
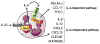
References
-
- Funakoshi-Tago M, Tago K, Hayakawa M, Tominaga S, Ohshio T, Sonoda Y, Kasahara T. TRAF6 is a critical signal transducer in IL-33 signaling pathway. Cell Signal. 2008;20:1679–1686. - PubMed
-
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. - PubMed
-
- Allakhverdi Z, Smith DE, Comeau MR, Delespesse G. Cutting edge: The ST2 ligand IL-33 potently activates and drives maturation of human mast cells. J Immunol. 2007;179:2051–2054. - PubMed
-
- Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, Pitman N, Mirchandani A, Rana B, van Rooijen N, Shepherd M, McSharry C, McInnes IB, Xu D, Liew FY. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–6477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

