Variant human T-cell lymphotropic virus type 1c and adult T-cell leukemia, Australia
- PMID: 24047544
- PMCID: PMC3810736
- DOI: 10.3201/eid1910.130105
Variant human T-cell lymphotropic virus type 1c and adult T-cell leukemia, Australia
Abstract
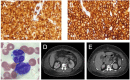
Human T-cell lymphotropic virus type 1 is endemic to central Australia among Indigenous Australians. However, virologic and clinical aspects of infection remain poorly understood. No attempt has been made to control transmission to indigenous children. We report 3 fatal cases of adult T-cell leukemia/lymphoma caused by human T-cell lymphotropic virus type 1 Australo-Melanesian subtype c.
Keywords: ATLL; Australia; HTLV-1; adult T-cell leukemia/lymphoma; human T-cell lymphotropic virus type 1; indigenous Australians; molecular epidemiology; viruses.
Figures
References
-
- May JT, Stent G, Schnagl RD. Antibody to human T-cell lymphotropic virus type I in Australian aborigines. Med J Aust. 1988;149:104 . - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
