Proliferative status regulates HDAC11 mRNA abundance in nontransformed fibroblasts
- PMID: 24047695
- PMCID: PMC3895431
- DOI: 10.4161/cc.26433
Proliferative status regulates HDAC11 mRNA abundance in nontransformed fibroblasts
Abstract
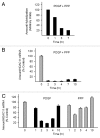
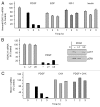
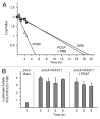
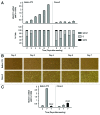
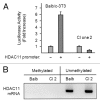
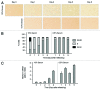
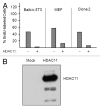
Histone deacetylases (HDACs) are important determinants of gene transcription and other biological processes. HDAC11 is one of the least characterized HDACs and is the only member of the class IV HDAC family. Our studies examined the events that control the expression of the HDAC11 transcript. We show that platelet-derived growth factor (PDGF) rapidly reduces the abundance of HDAC11 mRNA when added to density-arrested Balb/c-3T3 cells, which are nontransformed fibroblasts. Reduction required mRNA and protein synthesis, but not AKT or ERK activity, and resulted from accelerated turnover of the HDAC11 transcript. Reduction was transient in cells receiving PDGF alone but sustained in cells receiving both PDGF and platelet-poor plasma, which together promote G₀/G₁ traverse and S phase entry. Plasma alone did not appreciably reduce HDAC11 mRNA abundance, nor did epidermal growth factor, insulin-like growth factor, or insulin. HDAC11 mRNA accumulated in Balb/c-3T3 cells exiting the cell cycle due to density-dependent growth inhibition or serum deprivation. Of note, HDAC11 mRNA did not accumulate in a spontaneously transformed Balb/c-3T3 clonal variant (clone 2) that does not density arrest. The HDAC11 promoter was active in Balb/c-3T3 but not clone 2 cells; inactivity in clone 2 cells did not result from methylation of CpG islands. Overexpression of HDAC11 inhibited the cell cycle progression of both transformed and nontransformed fibroblasts. Our studies identify the HDAC11 transcript as a PDGF target and show that HDAC11 mRNA abundance correlates inversely with proliferative status.
Keywords: cell cycle; clone 2; fibroblasts; histone deacetylase 11; platelet-derived growth factor.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
