The estrogen receptor α is the key regulator of the bifunctional role of FoxO3a transcription factor in breast cancer motility and invasiveness
- PMID: 24047697
- PMCID: PMC3895429
- DOI: 10.4161/cc.26421
The estrogen receptor α is the key regulator of the bifunctional role of FoxO3a transcription factor in breast cancer motility and invasiveness
Abstract
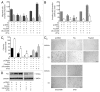
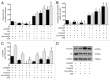
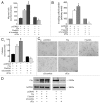
The role of the Forkhead box class O (FoxO)3a transcription factor in breast cancer migration and invasion is controversial. Here we show that FoxO3a overexpression decreases motility, invasiveness, and anchorage-independent growth in estrogen receptor α-positive (ERα+) cancer cells while eliciting opposite effects in ERα-silenced cells and in ERα-negative (ERα-) cell lines, demonstrating that the nuclear receptor represents a crucial switch in FoxO3a control of breast cancer cell aggressiveness. In ERα+ cells, FoxO3a-mediated events were paralleled by a significant induction of Caveolin-1 (Cav1), an essential constituent of caveolae negatively associated to tumor invasion and metastasis. Cav1 induction occurs at the transcriptional level through FoxO3a binding to a Forkhead responsive core sequence located at position -305/-299 of the Cav1 promoter. 17β-estradiol (E2) strongly emphasized FoxO3a effects on cell migration and invasion, while ERα and Cav1 silencing were able to reverse them, demonstrating that both proteins are pivotal mediators of these FoxO3a controlled processes. In vivo, an immunohistochemical analysis on tissue sections from patients with ERα+ or ERα- invasive breast cancers or in situ ductal carcinoma showed that nuclear FoxO3a inversely (ERα+) or directly (ERα-) correlated with the invasive phenotype of breast tumors. In conclusion, FoxO3a role in breast cancer motility and invasion depends on ERα status, disclosing a novel aspect of the well-established FoxO3a/ERα interplay. Therefore FoxO3a might become a pursuable target to be suitably exploited in combination therapies either in ERα+ or ERα- breast tumors.
Keywords: breast cancer; estrogen receptor; forkhead transcription factors; invasion; motility.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
