Complexes of Trypanosoma cruzi sterol 14α-demethylase (CYP51) with two pyridine-based drug candidates for Chagas disease: structural basis for pathogen selectivity
- PMID: 24047900
- PMCID: PMC3814756
- DOI: 10.1074/jbc.M113.497990
Complexes of Trypanosoma cruzi sterol 14α-demethylase (CYP51) with two pyridine-based drug candidates for Chagas disease: structural basis for pathogen selectivity
Abstract
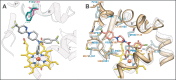
Chagas disease, caused by the eukaryotic (protozoan) parasite Trypanosoma cruzi, is an alarming emerging global health problem with no clinical drugs available to treat the chronic stage. Azole inhibitors of sterol 14α-demethylase (CYP51) were proven effective against Chagas, and antifungal drugs posaconazole and ravuconazole have entered clinical trials in Spain, Bolivia, and Argentina. Here we present the x-ray structures of T. cruzi CYP51 in complexes with two alternative drug candidates, pyridine derivatives (S)-(4-chlorophenyl)-1-(4-(4-(trifluoromethyl)phenyl)-piperazin-1-yl)-2-(pyridin-3-yl)ethanone (UDO; Protein Data Bank code 3ZG2) and N-[4-(trifluoromethyl)phenyl]-N-[1-[5-(trifluoromethyl)-2-pyridyl]-4-piperi-dyl]pyridin-3-amine (UDD; Protein Data Bank code 3ZG3). These compounds have been developed by the Drugs for Neglected Diseases initiative (DNDi) and are highly promising antichagasic agents in both cellular and in vivo experiments. The binding parameters and inhibitory effects on sterol 14α-demethylase activity in reconstituted enzyme reactions confirmed UDO and UDD as potent and selective T. cruzi CYP51 inhibitors. Comparative analysis of the pyridine- and azole-bound CYP51 structures uncovered the features that make UDO and UDD T. cruzi CYP51-specific. The structures suggest that although a precise fit between the shape of the inhibitor molecules and T. cruzi CYP51 active site topology underlies their high inhibitory potency, a longer coordination bond between the catalytic heme iron and the pyridine nitrogen implies a weaker influence of pyridines on the iron reduction potential, which may be the basis for the observed selectivity of these compounds toward the target enzyme versus other cytochrome P450s, including human drug-metabolizing P450s. These findings may pave the way for the development of novel CYP51-targeted drugs with optimized metabolic properties that are very much needed for the treatment of human infections caused by eukaryotic microbial pathogens.
Keywords: Cytochrome P450; Drug Discovery; Enzyme Inhibitors; Infectious Diseases; Protein Complexes; Protein Structure; Sterol 14α-Demethylase; Trypanosoma cruzi; X-ray Crystallography.
Figures





Similar articles
-
Development of a Fluorescence-based Trypanosoma cruzi CYP51 Inhibition Assay for Effective Compound Triaging in Drug Discovery Programmes for Chagas Disease.PLoS Negl Trop Dis. 2015 Sep 22;9(9):e0004014. doi: 10.1371/journal.pntd.0004014. eCollection 2015 Sep. PLoS Negl Trop Dis. 2015. PMID: 26394211 Free PMC article.
-
In vitro and in vivo studies of the antiparasitic activity of sterol 14α-demethylase (CYP51) inhibitor VNI against drug-resistant strains of Trypanosoma cruzi.Antimicrob Agents Chemother. 2013 Sep;57(9):4151-63. doi: 10.1128/AAC.00070-13. Epub 2013 Jun 17. Antimicrob Agents Chemother. 2013. PMID: 23774435 Free PMC article.
-
Clinical Candidate VT-1161's Antiparasitic Effect In Vitro, Activity in a Murine Model of Chagas Disease, and Structural Characterization in Complex with the Target Enzyme CYP51 from Trypanosoma cruzi.Antimicrob Agents Chemother. 2015 Dec 7;60(2):1058-66. doi: 10.1128/AAC.02287-15. Print 2016 Feb. Antimicrob Agents Chemother. 2015. PMID: 26643331 Free PMC article.
-
Sterol 14alpha-demethylase (CYP51) as a therapeutic target for human trypanosomiasis and leishmaniasis.Curr Top Med Chem. 2011;11(16):2060-71. doi: 10.2174/156802611796575902. Curr Top Med Chem. 2011. PMID: 21619513 Free PMC article. Review.
-
Design or screening of drugs for the treatment of Chagas disease: what shows the most promise?Expert Opin Drug Discov. 2013 Dec;8(12):1479-89. doi: 10.1517/17460441.2013.845554. Epub 2013 Sep 30. Expert Opin Drug Discov. 2013. PMID: 24079515 Free PMC article. Review.
Cited by
-
Novel structural CYP51 mutation in Trypanosoma cruzi associated with multidrug resistance to CYP51 inhibitors and reduced infectivity.Int J Parasitol Drugs Drug Resist. 2020 Aug;13:107-120. doi: 10.1016/j.ijpddr.2020.06.001. Epub 2020 Jun 13. Int J Parasitol Drugs Drug Resist. 2020. PMID: 32688218 Free PMC article.
-
The Evolution of Azole Resistance in Candida albicans Sterol 14α-Demethylase (CYP51) through Incremental Amino Acid Substitutions.Antimicrob Agents Chemother. 2019 Apr 25;63(5):e02586-18. doi: 10.1128/AAC.02586-18. Print 2019 May. Antimicrob Agents Chemother. 2019. PMID: 30783005 Free PMC article.
-
Repositioned Drugs for Chagas Disease Unveiled via Structure-Based Drug Repositioning.Int J Mol Sci. 2020 Nov 20;21(22):8809. doi: 10.3390/ijms21228809. Int J Mol Sci. 2020. PMID: 33233837 Free PMC article.
-
Development of a Fluorescence-based Trypanosoma cruzi CYP51 Inhibition Assay for Effective Compound Triaging in Drug Discovery Programmes for Chagas Disease.PLoS Negl Trop Dis. 2015 Sep 22;9(9):e0004014. doi: 10.1371/journal.pntd.0004014. eCollection 2015 Sep. PLoS Negl Trop Dis. 2015. PMID: 26394211 Free PMC article.
-
Crystal Structure of the New Investigational Drug Candidate VT-1598 in Complex with Aspergillus fumigatus Sterol 14α-Demethylase Provides Insights into Its Broad-Spectrum Antifungal Activity.Antimicrob Agents Chemother. 2017 Jun 27;61(7):e00570-17. doi: 10.1128/AAC.00570-17. Print 2017 Jul. Antimicrob Agents Chemother. 2017. PMID: 28461309 Free PMC article.
References
-
- Fischer R. T., Stam S. H., Johnson P. R., Ko S. S., Magolda R. L., Gaylor J. L., Trzaskos J. M. (1989) Mechanistic studies of lanosterol 14α-methyl demethylase: substrate requirements for the component reactions catalyzed by a single cytochrome P-450 isozyme. J. Lipid Res. 30, 1621–1632 - PubMed
-
- Trzaskos J. M., Bowen W. D., Shafiee A., Fischer R. T., Gaylor J. L. (1984) Cytochrome P-450-dependent oxidation of lanosterol in cholesterol biosynthesis. Microsomal electron transport and C-32 demethylation. J. Biol. Chem. 259, 13402–13412 - PubMed
-
- Lepesheva G. I., Waterman M. R. (2004) CYP51, the omnipotent P450. Mol. Cell. Endocrinol. 215, 165–170 - PubMed
-
- Nes W. R., McKean M. R. (eds) (1977) Biochemistry of Steroids and Other Isopentenoids, pp. 325–411, University Park Press, Baltimore
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical

