Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop
- PMID: 24048002
- PMCID: PMC3785887
- DOI: 10.1016/j.bpj.2013.07.049
Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop
Abstract
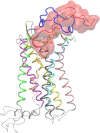
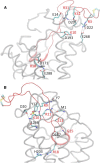
HIV-1 cell entry is initiated by the interaction of the viral envelope glycoprotein gp120 with CD4, and chemokine coreceptors CXCR4 and CCR5. The molecular recognition of CXCR4 or CCR5 by the HIV-1 gp120 is mediated through the V3 loop, a fragment of gp120. The binding of the V3 loop to CXCR4 or CCR5 determines the cell tropism of HIV-1 and constitutes a key step before HIV-1 cell entry. Thus, elucidating the molecular recognition of CXCR4 by the V3 loop is important for understanding HIV-1 viral infectivity and tropism, and for the design of HIV-1 inhibitors. We employed a comprehensive set of computational tools, predominantly based on free energy calculations and molecular-dynamics simulations, to investigate the molecular recognition of CXCR4 by a dual tropic V3 loop. We report what is, to our knowledge, the first HIV-1 gp120 V3 loop:CXCR4 complex structure. The computationally derived structure reveals an abundance of polar and nonpolar intermolecular interactions contributing to the HIV-1 gp120:CXCR4 binding. Our results are in remarkable agreement with previous experimental findings. Therefore, this work sheds light on the functional role of HIV-1 gp120 V3 loop and CXCR4 residues associated with HIV-1 coreceptor activity.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop.PLoS One. 2014 Apr 24;9(4):e95767. doi: 10.1371/journal.pone.0095767. eCollection 2014. PLoS One. 2014. PMID: 24763408 Free PMC article.
-
HIV-1 gp120-CXCR4 recognition probed with synthetic nanomolar affinity D-peptides containing fragments of gp120 V3 loop.Eur J Med Chem. 2022 Dec 15;244:114797. doi: 10.1016/j.ejmech.2022.114797. Epub 2022 Sep 28. Eur J Med Chem. 2022. PMID: 36270088 Free PMC article.
-
Role of the HIV type 1 glycoprotein 120 V3 loop in determining coreceptor usage.AIDS Res Hum Retroviruses. 1999 May 20;15(8):731-43. doi: 10.1089/088922299310827. AIDS Res Hum Retroviruses. 1999. PMID: 10357469
-
HIV-1 gp120 V3 loop for structure-based drug design.Curr Protein Pept Sci. 2005 Oct;6(5):413-22. doi: 10.2174/138920305774329359. Curr Protein Pept Sci. 2005. PMID: 16248793 Review.
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
Cited by
-
Unique profile of predominant CCR5-tropic in CRF07_BC HIV-1 infections and discovery of an unusual CXCR4-tropic strain.Front Immunol. 2022 Sep 23;13:911806. doi: 10.3389/fimmu.2022.911806. eCollection 2022. Front Immunol. 2022. PMID: 36211390 Free PMC article.
-
Function-oriented development of CXCR4 antagonists as selective human immunodeficiency virus (HIV)-1 entry inhibitors.J Med Chem. 2015 Feb 12;58(3):1452-65. doi: 10.1021/jm501772w. Epub 2015 Jan 28. J Med Chem. 2015. PMID: 25584630 Free PMC article.
-
Computational evolution of an RNA-binding protein towards enhanced oxidized-RNA binding.Comput Struct Biotechnol J. 2019 Dec 27;18:137-152. doi: 10.1016/j.csbj.2019.12.003. eCollection 2020. Comput Struct Biotechnol J. 2019. PMID: 31988703 Free PMC article.
-
Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop.PLoS One. 2014 Apr 24;9(4):e95767. doi: 10.1371/journal.pone.0095767. eCollection 2014. PLoS One. 2014. PMID: 24763408 Free PMC article.
-
Spermine and spermidine bind CXCR4 and inhibit CXCR4- but not CCR5-tropic HIV-1 infection.Sci Adv. 2023 Jul 7;9(27):eadf8251. doi: 10.1126/sciadv.adf8251. Epub 2023 Jul 5. Sci Adv. 2023. PMID: 37406129 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

