Dynamic transition states of ErbB1 phosphorylation predicted by spatial stochastic modeling
- PMID: 24048005
- PMCID: PMC3785885
- DOI: 10.1016/j.bpj.2013.07.056
Dynamic transition states of ErbB1 phosphorylation predicted by spatial stochastic modeling
Abstract
ErbB1 overexpression is strongly linked to carcinogenesis, motivating better understanding of erbB1 dimerization and activation. Recent single-particle-tracking data have provided improved measures of dimer lifetimes and strong evidence that transient receptor coconfinement promotes repeated interactions between erbB1 monomers. Here, spatial stochastic simulations explore the potential impact of these parameters on erbB1 phosphorylation kinetics. This rule-based mathematical model incorporates structural evidence for conformational flux of the erbB1 extracellular domains, as well as asymmetrical orientation of erbB1 cytoplasmic kinase domains during dimerization. The asymmetric dimer model considers the theoretical consequences of restricted transactivation of erbB1 receptors within a dimer, where the N-lobe of one monomer docks with the C-lobe of the second monomer and triggers its catalytic activity. The dynamic nature of the erbB1 phosphorylation state is shown by monitoring activation states of individual monomers as they diffuse, bind, and rebind after ligand addition. The model reveals the complex interplay between interacting liganded and nonliganded species and the influence of their distribution and abundance within features of the membrane landscape.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

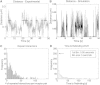
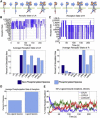
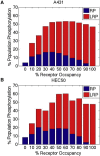

References
-
- Citri A., Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. - PubMed
-
- Baselga J., Swain S.M. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer. 2009;9:463–475. - PubMed
-
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. - PubMed
-
- Hsieh M.Y., Yang S., Edwards J.S. Stochastic simulations of ErbB homo and heterodimerisation: potential impacts of receptor conformational state and spatial segregation. IET Syst. Biol. 2008;2:256–272. - PubMed
-
- Sasagawa S., Ozaki Y., Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 2005;7:365–373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

