Bves and NDRG4 regulate directional epicardial cell migration through autocrine extracellular matrix deposition
- PMID: 24048452
- PMCID: PMC3826988
- DOI: 10.1091/mbc.E12-07-0539
Bves and NDRG4 regulate directional epicardial cell migration through autocrine extracellular matrix deposition
Abstract
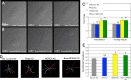
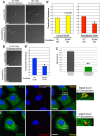
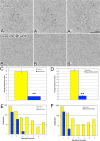
Directional cell movement is universally required for tissue morphogenesis. Although it is known that cell/matrix interactions are essential for directional movement in heart development, the mechanisms governing these interactions require elucidation. Here we demonstrate that a novel protein/protein interaction between blood vessel epicardial substance (Bves) and N-myc downstream regulated gene 4 (NDRG4) is critical for regulation of epicardial cell directional movement, as disruption of this interaction randomizes migratory patterns. Our studies show that Bves/NDRG4 interaction is required for trafficking of internalized fibronectin through the "autocrine extracellular matrix (ECM) deposition" fibronectin recycling pathway. Of importance, we demonstrate that Bves/NDRG4-mediated fibronectin recycling is indeed essential for epicardial cell directional movement, thus linking these two cell processes. Finally, total internal reflectance fluorescence microscopy shows that Bves/NDRG4 interaction is required for fusion of recycling endosomes with the basal cell surface, providing a molecular mechanism of motility substrate delivery that regulates cell directional movement. This is the first evidence of a molecular function for Bves and NDRG4 proteins within broader subcellular trafficking paradigms. These data identify novel regulators of a critical vesicle-docking step required for autocrine ECM deposition and explain how Bves facilitates cell-microenvironment interactions in the regulation of epicardial cell-directed movement.
Figures

Similar articles
-
Identification of a novel Bves function: regulation of vesicular transport.EMBO J. 2010 Feb 3;29(3):532-45. doi: 10.1038/emboj.2009.379. Epub 2010 Jan 7. EMBO J. 2010. PMID: 20057356 Free PMC article.
-
Membrane topology of Bves/Pop1A, a cell adhesion molecule that displays dynamic changes in cellular distribution during development.J Biol Chem. 2003 Aug 29;278(35):32872-9. doi: 10.1074/jbc.M301961200. Epub 2003 Jun 17. J Biol Chem. 2003. PMID: 12815060
-
Bves modulates epithelial integrity through an interaction at the tight junction.J Cell Sci. 2005 Oct 15;118(Pt 20):4667-78. doi: 10.1242/jcs.02588. Epub 2005 Sep 27. J Cell Sci. 2005. PMID: 16188940
-
Bves, a member of the Popeye domain-containing gene family.Dev Dyn. 2006 Mar;235(3):586-93. doi: 10.1002/dvdy.20688. Dev Dyn. 2006. PMID: 16444674 Free PMC article. Review.
-
Blood Vessel Epicardial Substance (BVES) in junctional signaling and cancer.Tissue Barriers. 2018;6(4):1-12. doi: 10.1080/21688370.2018.1499843. Epub 2018 Oct 11. Tissue Barriers. 2018. PMID: 30307367 Free PMC article. Review.
Cited by
-
The cAMP-binding Popdc proteins have a redundant function in the heart.Biochem Soc Trans. 2014 Apr;42(2):295-301. doi: 10.1042/BST20130264. Biochem Soc Trans. 2014. PMID: 24646234 Free PMC article. Review.
-
Collaboration of fibronectin matrix with other extracellular signals in morphogenesis and differentiation.Curr Opin Cell Biol. 2016 Oct;42:1-6. doi: 10.1016/j.ceb.2016.03.014. Epub 2016 Apr 7. Curr Opin Cell Biol. 2016. PMID: 27062478 Free PMC article. Review.
-
An interaction of heart disease-associated proteins POPDC1/2 with XIRP1 in transverse tubules and intercalated discs.BMC Mol Cell Biol. 2020 Dec 1;21(1):88. doi: 10.1186/s12860-020-00329-3. BMC Mol Cell Biol. 2020. PMID: 33261556 Free PMC article.
-
A loss-of-function mutation p.T256M in NDRG4 is implicated in the pathogenesis of pulmonary atresia with ventricular septal defect (PA/VSD) and tetralogy of Fallot (TOF).FEBS Open Bio. 2021 Feb;11(2):375-385. doi: 10.1002/2211-5463.13044. Epub 2021 Jan 9. FEBS Open Bio. 2021. PMID: 33211401 Free PMC article.
-
Neuronal Ndrg4 Is Essential for Nodes of Ranvier Organization in Zebrafish.PLoS Genet. 2016 Nov 30;12(11):e1006459. doi: 10.1371/journal.pgen.1006459. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27902705 Free PMC article.
References
-
- Andrée B, Hillemann T, Kessler-Icekson G, Schmitt-John T, Jockusch H, Arnold HH, Brand T. Isolation and characterization of the novel Popeye gene family expressed in skeletal muscle and heart. Dev Biol. 2000;223:371–382. - PubMed
-
- Berger P, Niemann A, Suter U. Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease) Glia. 2006;54:243–257. - PubMed
-
- Choi S-C, et al. NDRG2 is one of novel intrinsic factors for regulation of IL-10 production in human myeloid cell. Biochem Biophys Res Commun. 2010;396:684–690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

