A host-based RT-PCR gene expression signature to identify acute respiratory viral infection
- PMID: 24048524
- PMCID: PMC4286889
- DOI: 10.1126/scitranslmed.3006280
A host-based RT-PCR gene expression signature to identify acute respiratory viral infection
Abstract
Improved ways to diagnose acute respiratory viral infections could decrease inappropriate antibacterial use and serve as a vital triage mechanism in the event of a potential viral pandemic. Measurement of the host response to infection is an alternative to pathogen-based diagnostic testing and may improve diagnostic accuracy. We have developed a host-based assay with a reverse transcription polymerase chain reaction (RT-PCR) TaqMan low-density array (TLDA) platform for classifying respiratory viral infection. We developed the assay using two cohorts experimentally infected with influenza A H3N2/Wisconsin or influenza A H1N1/Brisbane, and validated the assay in a sample of adults presenting to the emergency department with fever (n = 102) and in healthy volunteers (n = 41). Peripheral blood RNA samples were obtained from individuals who underwent experimental viral challenge or who presented to the emergency department and had microbiologically proven viral respiratory infection or systemic bacterial infection. The selected gene set on the RT-PCR TLDA assay classified participants with experimentally induced influenza H3N2 and H1N1 infection with 100 and 87% accuracy, respectively. We validated this host gene expression signature in a cohort of 102 individuals arriving at the emergency department. The sensitivity of the RT-PCR test was 89% [95% confidence interval (CI), 72 to 98%], and the specificity was 94% (95% CI, 86 to 99%). These results show that RT-PCR-based detection of a host gene expression signature can classify individuals with respiratory viral infection and sets the stage for prospective evaluation of this diagnostic approach in a clinical setting.
Conflict of interest statement
Figures
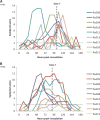


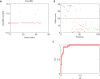
References
-
- Silvennoinen H, Peltola V, Vainionpää R, Ruuskanen O, Heikkinen T. Incidence of influenza-related hospitalizations in different age groups of children in Finland: A 16-year study. Pediatr Infect Dis J. 2011;30:e24–e28. - PubMed
-
- Apisarnthanarak A, Mundy LM. Rapid testing for pandemic influenza A (H1N1): Diagnostic test utility and specimen source. Infect Control Hosp Epidemiol. 2010;31:663–664. - PubMed
-
- Ciblak MA, Kanturvardar M, Asar S, Bozkaya E, Yenen OS, Badur S. Sensitivity of rapid influenza antigen tests in the diagnosis of pandemic (H1N1)2009 compared with the standard rRT-PCR technique during the 2009 pandemic in Turkey. Scand J Infect Dis. 2010;42:902–905. - PubMed
-
- Cunha BA, Syed U, Stroll S, Mickail N, Laguerre M. Winthrop-University Hospital Infectious Disease Division’s swine influenza (H1N1) pneumonia diagnostic weighted point score system for hospitalized adults with influenza-like illnesses (ILIs) and negative rapid influenza diagnostic tests (RIDTs) Heart Lung. 2009;38:534–538. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

