Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline
- PMID: 24048575
- PMCID: PMC4076377
- DOI: 10.1095/biolreprod.113.113308
Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline
Abstract
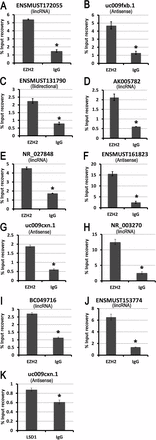
In mammals, the transcriptome of large noncoding RNAs (lncRNAs) is believed to be greater than that of messenger RNAs (mRNAs). Some lncRNAs, especially large intergenic noncoding RNAs (lincRNAs), participate in epigenetic regulation by binding chromatin-modifying protein complexes and regulating protein-coding gene expression. Given that epigenetic regulation plays a critical role in male germline development, we embarked on expression profiling of both lncRNAs and mRNAs during male germline reprogramming and postnatal development using microarray analyses. We identified thousands of lncRNAs and hundreds of lincRNAs that are either up- or downregulated at six critical time points during male germ cell development. In addition, highly regulated lncRNAs were correlated with nearby (<30 kb) mRNA gene clusters, which were also significantly up- or downregulated. Large ncRNAs can be localized to both the nucleus and cytoplasm, with nuclear lncRNAs mostly associated with key components of the chromatin-remodeling protein complexes. Our data indicate that expression of lncRNAs is dynamically regulated during male germline development and that lncRNAs may function to regulate gene expression at both transcriptional and posttranscriptional levels via genetic and epigenetic mechanisms.
Keywords: epigenetics; fertility; germ cell; reproduction; spermatogenesis.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

