Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase
- PMID: 24048591
- PMCID: PMC3809364
- DOI: 10.1242/dev.096081
Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase
Abstract
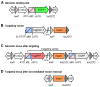
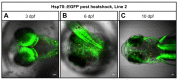
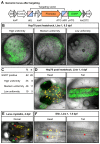
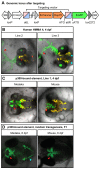
Established transgenesis methods for fish model systems allow efficient genomic integration of transgenes. However, thus far a way of controlling copy number and integration sites has not been available, leading to variable transgene expression caused by position effects. The integration of transgenes at predefined genomic positions enables the direct comparison of different transgenes, thereby improving time and cost efficiency. Here, we report an efficient PhiC31-based site-specific transgenesis system for medaka. This system includes features that allow the pre-selection of successfully targeted integrations early on in the injected generation. Pre-selected embryos transmit the correctly integrated transgene through the germline with high efficiency. The landing site design enables a variety of applications, such as reporter and enhancer switch, in addition to the integration of any insert. Importantly, this allows assaying of enhancer activity in a site-specific manner without requiring germline transmission, thus speeding up large-scale analyses of regulatory elements.
Keywords: Fish; In vivo analysis; PhiC31 integrase; Position effects; Regulatory DNA.
Figures


References
-
- Barolo S. (2006). Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25, 7505–7511 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

