Role of a ubiquitously expressed receptor in the vertebrate olfactory system
- PMID: 24048853
- PMCID: PMC3776066
- DOI: 10.1523/JNEUROSCI.2339-13.2013
Role of a ubiquitously expressed receptor in the vertebrate olfactory system
Abstract
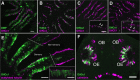
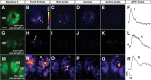
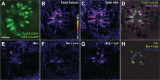
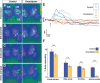
Odorant cues are recognized by receptors expressed on olfactory sensory neurons, the primary sensory neurons of the olfactory epithelium. Odorant receptors typically obey the "one receptor, one neuron" rule, in which the receptive field of the olfactory neuron is determined by the singular odorant receptor that it expresses. Odor-evoked receptor activity across the population of olfactory neurons is then interpreted by the brain to identify the molecular nature of the odorant stimulus. In the present study, we characterized the properties of a C family G-protein-coupled receptor that, unlike most other odorant receptors, is expressed in a large population of microvillous sensory neurons in the zebrafish olfactory epithelium and the mouse vomeronasal organ. We found that this receptor, OlfCc1 in zebrafish and its murine ortholog Vmn2r1, is a calcium-dependent, low-sensitivity receptor specific for the hydrophobic amino acids isoleucine, leucine, and valine. Loss-of-function experiments in zebrafish embryos demonstrate that OlfCc1 is required for olfactory responses to a diverse mixture of polar, nonpolar, acidic, and basic amino acids. OlfCc1 was also found to promote localization of other OlfC receptor family members to the plasma membrane in heterologous cells. Together, these results suggest that the broadly expressed OlfCc1 is required for amino acid detection by the olfactory system and suggest that it plays a role in the function and/or intracellular trafficking of other olfactory and vomeronasal receptors with which it is coexpressed.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
