Dynamics of viral evolution and neutralizing antibody response after HIV-1 superinfection
- PMID: 24049166
- PMCID: PMC3838120
- DOI: 10.1128/JVI.02260-13
Dynamics of viral evolution and neutralizing antibody response after HIV-1 superinfection
Abstract
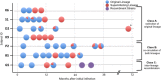
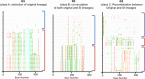
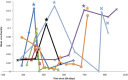
Investigating the incidence and prevalence of HIV-1 superinfection is challenging due to the complex dynamics of two infecting strains. The superinfecting strain can replace the initial strain, be transiently expressed, or persist along with the initial strain in distinct or in recombined forms. Various selective pressures influence these alternative scenarios in different HIV-1 coding regions. We hypothesized that the potency of the neutralizing antibody (NAb) response to autologous viruses would modulate viral dynamics in env following superinfection in a limited set of superinfection cases. HIV-1 env pyrosequencing data were generated from blood plasma collected from 7 individuals with evidence of superinfection. Viral variants within each patient were screened for recombination, and viral dynamics were evaluated using nucleotide diversity. NAb responses to autologous viruses were evaluated before and after superinfection. In 4 individuals, the superinfecting strain replaced the original strain. In 2 individuals, both initial and superinfecting strains continued to cocirculate. In the final individual, the surviving lineage was the product of interstrain recombination. NAb responses to autologous viruses that were detected within the first 2 years of HIV-1 infection were weak or absent for 6 of the 7 recently infected individuals at the time of and shortly following superinfection. These 6 individuals had detectable on-going viral replication of distinct superinfecting virus in the env coding region. In the remaining case, there was an early and strong autologous NAb response, which was associated with extensive recombination in env between initial and superinfecting strains. This extensive recombination made superinfection more difficult to identify and may explain why the detection of superinfection has typically been associated with low autologous NAb titers.
Figures

References
-
- Ramos A, Hu DJ, Nguyen L, Phan K-O, Vanichseni S, Promadej N, Choopanya K, Callahan M, Young NL, McNicholl J, Mastro TD, Folks TM, Subbarao S. 2002. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 76:7444–7452 - PMC - PubMed
-
- Jost S, Bernard MC, Kaiser L, Yerly S, Hirschel B, Samri A, Autran B, Goh LE, Perrin L. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347:731–736 - PubMed
-
- Smith DM, Wong JK, Hightower GK, Ignacio CC, Koelsch KK, Petropoulos CJ, Richman DD, Little SJ. 2005. HIV drug resistance acquired through superinfection. AIDS 19:1251–1256 - PubMed
-
- Gottlieb GS, Nickle DC, Jensen MA, Wong KG, Grobler J, Li F, Liu S-L, Rademeyer C, Learn GH, Karim SSA, Williamson C, Corey L, Margolick JB, Mullins JI. 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363:619–622 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K24 AI100665/AI/NIAID NIH HHS/United States
- AI43629/AI/NIAID NIH HHS/United States
- R01 AI047745/AI/NIAID NIH HHS/United States
- AI36214/AI/NIAID NIH HHS/United States
- AI74621/AI/NIAID NIH HHS/United States
- DA034978/DA/NIDA NIH HHS/United States
- R01 MH097520/MH/NIMH NIH HHS/United States
- AI090970/AI/NIAID NIH HHS/United States
- P01 AI074621/AI/NIAID NIH HHS/United States
- R24 AI106039/AI/NIAID NIH HHS/United States
- MH097520/MH/NIMH NIH HHS/United States
- R21 AI047745/AI/NIAID NIH HHS/United States
- T32 AI007384/AI/NIAID NIH HHS/United States
- DP1 DA034978/DA/NIDA NIH HHS/United States
- R56 AI047745/AI/NIAID NIH HHS/United States
- R01 AI043629/AI/NIAID NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- AI100665/AI/NIAID NIH HHS/United States
- U19 AI090970/AI/NIAID NIH HHS/United States
- AI47745/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

