Epstein-Barr virus and human herpesvirus 6 detection in a non-Hodgkin's diffuse large B-cell lymphoma cohort by using RNA sequencing
- PMID: 24049168
- PMCID: PMC3838131
- DOI: 10.1128/JVI.02380-13
Epstein-Barr virus and human herpesvirus 6 detection in a non-Hodgkin's diffuse large B-cell lymphoma cohort by using RNA sequencing
Erratum in
-
Correction for Strong et al., Epstein-Barr Virus and Human Herpesvirus 6 Detection in a Non-Hodgkin's Diffuse Large B-Cell Lymphoma Cohort by Using RNA Sequencing.J Virol. 2015 Dec 30;90(2):1152. doi: 10.1128/JVI.02772-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26719559 Free PMC article. No abstract available.
Abstract
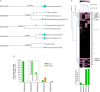
Comprehensive virome analysis of RNA sequence (RNA-seq) data sets from 118 non-Hodgkin's B-cell lymphomas revealed a small subset that is positive for Epstein-Barr virus (EBV) or human herpesvirus 6B (HHV-6B), with one coinfection. EBV transcriptome analysis revealed expression of the latency genes RPMS1, LMP1, and LMP2, with one sample additionally showing a high level of early lytic expression and another sample showing a high level of EBNA2 expression. HHV-6B transcriptome analysis revealed that the majority of genes were transcribed.
Figures

References
-
- Ogata M. 2009. Human herpesvirus 6 in hematological malignancies. J. Clin. Exp. Hematopathol. 49:57–67 - PubMed
-
- Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, Strong AL, Lehman TA, Seddon MB, Lin Z, Concha M, Baddoo M, Ferris M, Swan KF, Sullivan DE, Burow ME, Taylor CM, Flemington EK. 2013. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog. 9:e1003341.10.1371/journal.ppat.1003341 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

