Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration
- PMID: 24049186
- PMCID: PMC3838128
- DOI: 10.1128/JVI.01942-13
Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration
Erratum in
-
Correction for Gupta et al., "Bromo- and Extraterminal Domain Chromatin Regulators Serve as Cofactors for Murine Leukemia Virus Integration".J Virol. 2024 Apr 16;98(4):e0013524. doi: 10.1128/jvi.00135-24. Epub 2024 Mar 13. J Virol. 2024. PMID: 38477531 Free PMC article. No abstract available.
Abstract
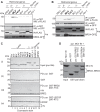
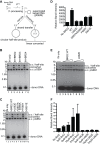
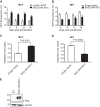
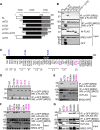
Retroviral integrase (IN) proteins catalyze the permanent integration of proviral genomes into host DNA with the help of cellular cofactors. Lens epithelium-derived growth factor (LEDGF) is a cofactor for lentiviruses, including human immunodeficiency virus type 1 (HIV-1), and targets lentiviral integration toward active transcription units in the host genome. In contrast to lentiviruses, murine leukemia virus (MLV), a gammaretrovirus, tends to integrate near transcription start sites. Here, we show that the bromodomain and extraterminal domain (BET) proteins BRD2, BRD3, and BRD4 interact with gammaretroviral INs and stimulate the catalytic activity of MLV IN in vitro. We mapped the interaction site to a characteristic structural feature within the BET protein extraterminal (ET) domain and to three amino acids in MLV IN. The ET domains of different BET proteins stimulate MLV integration in vitro and, in the case of BRD2, also in vivo. Furthermore, two small-molecule BET inhibitors, JQ1 and I-BET, decrease MLV integration and shift it away from transcription start sites. Our data suggest that BET proteins might act as chromatin-bound acceptors for the MLV preintegration complex. These results could pave a way to redirecting MLV DNA integration as a basis for creating safer retroviral vectors.
Figures




References
-
- Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

