Intraspecific variation of recombination rate in maize
- PMID: 24050704
- PMCID: PMC4053771
- DOI: 10.1186/gb-2013-14-9-r103
Intraspecific variation of recombination rate in maize
Abstract
Background: In sexually reproducing organisms, meiotic crossovers ensure the proper segregation of chromosomes and contribute to genetic diversity by shuffling allelic combinations. Such genetic reassortment is exploited in breeding to combine favorable alleles, and in genetic research to identify genetic factors underlying traits of interest via linkage or association-based approaches. Crossover numbers and distributions along chromosomes vary between species, but little is known about their intraspecies variation.
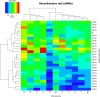
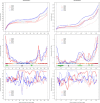
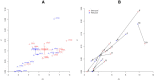
Results: Here, we report on the variation of recombination rates between 22 European maize inbred lines that belong to the Dent and Flint gene pools. We genotype 23 doubled-haploid populations derived from crosses between these lines with a 50 k-SNP array and construct high-density genetic maps, showing good correspondence with the maize B73 genome sequence assembly. By aligning each genetic map to the B73 sequence, we obtain the recombination rates along chromosomes specific to each population. We identify significant differences in recombination rates at the genome-wide, chromosome, and intrachromosomal levels between populations, as well as significant variation for genome-wide recombination rates among maize lines. Crossover interference analysis using a two-pathway modeling framework reveals a negative association between re combination rate and interference strength.
Conclusions: To our knowledge, the present work provides the most comprehensive study on intraspecific variation of recombination rates and crossover interference strength in eukaryotes. Differences found in recombination rates will allow for selection of high or low recombining lines in crossing programs. Our methodology should pave the way for precise identification of genes controlling recombination rates in maize and other organisms.
Figures

References
-
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci. 2002;14:1611–1622. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

