Ligand-induced stabilization of the aptamer terminal helix in the add adenine riboswitch
- PMID: 24051105
- PMCID: PMC3851719
- DOI: 10.1261/rna.040493.113
Ligand-induced stabilization of the aptamer terminal helix in the add adenine riboswitch
Abstract
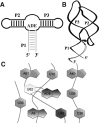
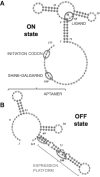
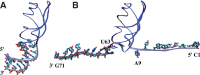
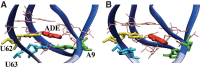
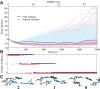
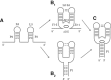
Riboswitches are structured mRNA elements that modulate gene expression. They undergo conformational changes triggered by highly specific interactions with sensed metabolites. Among the structural rearrangements engaged by riboswitches, the forming and melting of the aptamer terminal helix, the so-called P1 stem, is essential for genetic control. The structural mechanisms by which this conformational change is modulated upon ligand binding mostly remain to be elucidated. Here, we used pulling molecular dynamics simulations to study the thermodynamics of the P1 stem in the add adenine riboswitch. The P1 ligand-dependent stabilization was quantified in terms of free energy and compared with thermodynamic data. This comparison suggests a model for the aptamer folding in which direct P1-ligand interactions play a minor role on the conformational switch when compared with those related to the ligand-induced aptamer preorganization.
Keywords: P1 stem; RNA aptamer; adenine riboswitch; free energy calculation; molecular dynamics simulation.
Figures


References
-
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 - PubMed
-
- Bayly CI, Cieplak P, Cornell W, Kollman PA 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J Phys Chem 97: 10269–10280
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
