Microbiota impact on the epigenetic regulation of colorectal cancer
- PMID: 24051204
- PMCID: PMC3851951
- DOI: 10.1016/j.molmed.2013.08.005
Microbiota impact on the epigenetic regulation of colorectal cancer
Abstract
Mechanisms of colorectal cancer (CRC) development can be generally divided into three categories: genetic, epigenetic, and aberrant immunologic signaling pathways, all of which may be triggered by an imbalanced intestinal microbiota. Aberrant gut microbial composition, termed 'dysbiosis', has been reported in inflammatory bowel disease patients who are at increased risk for CRC development. Recent studies indicate that it is feasible to rescue experimental models of colonic cancer by oral treatment with genetically engineered beneficial bacteria and/or their immune-regulating gene products. Here, we review the mechanisms of epigenetic modulation implicated in the development and progression of CRC, which may be the result of dysbiosis, and therefore may be amenable to therapeutic intervention.
Keywords: colorectal cancer; commensal bacteria; epigenetic regulation; inflammatory bowel disease; microbiota.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


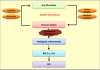
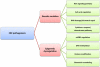

References
-
- Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
-
- Tjalsma H, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. - PubMed
-
- Tudek B, Speina E. Oxidatively damaged DNA and its repair in colon carcinogenesis. Mutat Res. 2012;736:82–92. - PubMed
-
- Arends MJ. Pathways of colorectal carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:97–102. - PubMed
-
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

