Neutrophil elastase and proteinase-3 trigger G protein-biased signaling through proteinase-activated receptor-1 (PAR1)
- PMID: 24052258
- PMCID: PMC3829148
- DOI: 10.1074/jbc.M113.483123
Neutrophil elastase and proteinase-3 trigger G protein-biased signaling through proteinase-activated receptor-1 (PAR1)
Abstract
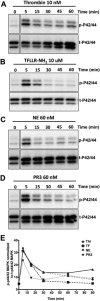
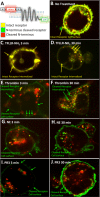
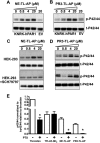
Neutrophil proteinases released at sites of inflammation can affect tissue function by either activating or disarming signal transduction mediated by proteinase-activated receptors (PARs). Because PAR1 is expressed at sites where abundant neutrophil infiltration occurs, we hypothesized that neutrophil-derived enzymes might also regulate PAR1 signaling. We report here that both neutrophil elastase and proteinase-3 cleave the human PAR1 N terminus at sites distinct from the thrombin cleavage site. This cleavage results in a disarming of thrombin-activated calcium signaling through PAR1. However, the distinct non-canonical tethered ligands unmasked by neutrophil elastase and proteinase-3, as well as synthetic peptides with sequences derived from these novel exposed tethered ligands, selectively stimulated PAR1-mediated mitogen-activated protein kinase activation. This signaling was blocked by pertussis toxin, implicating a Gαi-triggered signal pathway. We conclude that neutrophil proteinases trigger biased PAR1 signaling and we describe a novel set of tethered ligands that are distinct from the classical tethered ligand revealed by thrombin. We further demonstrate the function of this biased signaling in regulating endothelial cell barrier integrity.
Keywords: Biased Signaling; Calcium Signaling; Endothelial Cell; G Protein-coupled Receptors (GPCR); Intracellular Trafficking; MAP Kinases (MAPKs); Neutrophil; Proteinase-activated Receptor; Thrombin.
Figures





Similar articles
-
Neutrophil elastase acts as a biased agonist for proteinase-activated receptor-2 (PAR2).J Biol Chem. 2011 Jul 15;286(28):24638-48. doi: 10.1074/jbc.M110.201988. Epub 2011 May 16. J Biol Chem. 2011. PMID: 21576245 Free PMC article.
-
Molecular basis for activation and biased signaling at the thrombin-activated GPCR proteinase activated receptor-4 (PAR4).J Biol Chem. 2020 Feb 21;295(8):2520-2540. doi: 10.1074/jbc.RA119.011461. Epub 2019 Dec 31. J Biol Chem. 2020. PMID: 31892516 Free PMC article.
-
PR3 and elastase alter PAR1 signaling and trigger vWF release via a calcium-independent mechanism from glomerular endothelial cells.PLoS One. 2012;7(8):e43916. doi: 10.1371/journal.pone.0043916. Epub 2012 Aug 29. PLoS One. 2012. PMID: 22952809 Free PMC article.
-
A New Concept of Action of Hemostatic Proteases on Inflammation, Neurotoxicity, and Tissue Regeneration.Biochemistry (Mosc). 2017 Jul;82(7):778-790. doi: 10.1134/S0006297917070033. Biochemistry (Mosc). 2017. PMID: 28918742 Review.
-
The domino effect triggered by the tethered ligand of the protease activated receptors.Thromb Res. 2020 Dec;196:87-98. doi: 10.1016/j.thromres.2020.08.004. Epub 2020 Aug 4. Thromb Res. 2020. PMID: 32853981 Free PMC article. Review.
Cited by
-
Three Diseases Mediated by Different Immunopathologic Mechanisms-ANCA-Associated Vasculitis, Anti-Glomerular Basement Membrane Disease, and Immune Complex-Mediated Glomerulonephritis-A Common Clinical and Histopathologic Picture: Rapidly Progressive Crescentic Glomerulonephritis.Biomedicines. 2023 Nov 6;11(11):2978. doi: 10.3390/biomedicines11112978. Biomedicines. 2023. PMID: 38001978 Free PMC article. Review.
-
Neutrophil Elastase Subverts the Immune Response by Cleaving Toll-Like Receptors and Cytokines in Pneumococcal Pneumonia.Front Immunol. 2018 Apr 25;9:732. doi: 10.3389/fimmu.2018.00732. eCollection 2018. Front Immunol. 2018. PMID: 29922273 Free PMC article.
-
Cathepsin S causes inflammatory pain via biased agonism of PAR2 and TRPV4.J Biol Chem. 2014 Sep 26;289(39):27215-27234. doi: 10.1074/jbc.M114.599712. Epub 2014 Aug 12. J Biol Chem. 2014. PMID: 25118282 Free PMC article.
-
Integrating the GPCR transactivation-dependent and biased signalling paradigms in the context of PAR1 signalling.Br J Pharmacol. 2016 Oct;173(20):2992-3000. doi: 10.1111/bph.13398. Epub 2016 Feb 16. Br J Pharmacol. 2016. PMID: 26624252 Free PMC article. Review.
-
Biased signalling and proteinase-activated receptors (PARs): targeting inflammatory disease.Br J Pharmacol. 2014 Mar;171(5):1180-94. doi: 10.1111/bph.12544. Br J Pharmacol. 2014. PMID: 24354792 Free PMC article. Review.
References
-
- Adams M. N., Ramachandran R., Yau M. K., Suen J. Y., Fairlie D. P., Hollenberg M. D., Hooper J. D. (2011) Structure, function and pathophysiology of protease activated receptors. Pharmacol. Ther. 130, 248–282 - PubMed
-
- Rasmussen U. B., Vouret-Craviari V., Jallat S., Schlesinger Y., Pagès G., Pavirani A., Lecocq J. P., Pouysségur J., Van Obberghen-Schilling E. (1991) cDNA cloning and expression of a hamster α-thrombin receptor coupled to Ca2+ mobilization. FEBS Lett. 288, 123–128 - PubMed
-
- Vu T. K., Hung D. T., Wheaton V. I., Coughlin S. R. (1991) Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 64, 1057–1068 - PubMed
-
- Vu T. K., Wheaton V. I., Hung D. T., Charo I., Coughlin S. R. (1991) Domains specifying thrombin-receptor interaction. Nature 353, 674–677 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

