CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: its selective dependence on IL-34
- PMID: 24052571
- PMCID: PMC3868188
- DOI: 10.1189/jlb.0613311
CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: its selective dependence on IL-34
Abstract
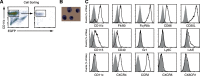
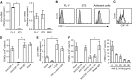
With the use of a mouse FDC line, FL-Y, we have been analyzing roles for FDCs in controlling B cell fate in GCs. Beside these regulatory functions, we fortuitously found that FL-Y cells induced a new type of CD11b⁺ monocytic cells (F4/80⁺, Gr-1⁻, Ly6C⁻, I-A/E(-/lo), CD11c⁻, CD115⁺, CXCR4⁺, CCR2⁺, CX₃CR1⁻) when cultured with a Lin⁻c-kit⁺ population from mouse spleen cells. The developed CD11b⁺ cells shared a similar gene-expression profile to mononuclear phagocytes and were designated as FDMCs. Here, we describe characteristic immunological functions and the induction mechanism of FDMCs. Proliferation of anti-CD40 antibody-stimulated B cells was markedly accelerated in the presence of FDMCs. In addition, the FDMC-activated B cells efficiently acquired GC B cell-associated markers (Fas and GL-7). We observed an increase of FDMC-like cells in mice after immunization. On the other hand, FL-Y cells were found to produce CSF-1 as well as IL-34, both of which are known to induce development of macrophages and monocytes by binding to the common receptor, CSF-1R, expressed on the progenitors. However, we show that FL-Y-derived IL-34, but not CSF-1, was selectively responsible for FDMC generation using neutralizing antibodies and RNAi. We also confirmed that FDMC generation was strictly dependent on CSF-1R. To our knowledge, a CSF-1R-mediated differentiation process that is intrinsically specific for IL-34 has not been reported. Our results provide new insights into understanding the diversity of IL-34 and CSF-1 signaling pathways through CSF-1R.
Keywords: CD11b; follicular dendritic cells; mouse spleen.
Figures






Comment in
-
The emerging specificities of interleukin-34.J Leukoc Biol. 2014 Jan;95(1):3-5. doi: 10.1189/jlb.0813466. J Leukoc Biol. 2014. PMID: 24385576 No abstract available.
Similar articles
-
Interleukin 34 (IL-34) cell-surface localization regulated by the molecular chaperone 78-kDa glucose-regulated protein facilitates the differentiation of monocytic cells.J Biol Chem. 2019 Feb 15;294(7):2386-2396. doi: 10.1074/jbc.RA118.006226. Epub 2018 Dec 20. J Biol Chem. 2019. PMID: 30573681 Free PMC article.
-
The emerging specificities of interleukin-34.J Leukoc Biol. 2014 Jan;95(1):3-5. doi: 10.1189/jlb.0813466. J Leukoc Biol. 2014. PMID: 24385576 No abstract available.
-
The immunoreceptor SLAMF8 promotes the differentiation of follicular dendritic cell-dependent monocytic cells with B cell-activating ability.FEBS Lett. 2022 Oct;596(20):2659-2667. doi: 10.1002/1873-3468.14468. Epub 2022 Aug 19. FEBS Lett. 2022. PMID: 35953458
-
Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells.Leukemia. 1996 Apr;10(4):588-99. Leukemia. 1996. PMID: 8618433 Review.
-
Regulation of cell cycle entry and G1 progression by CSF-1.Mol Reprod Dev. 1997 Jan;46(1):11-8. doi: 10.1002/(SICI)1098-2795(199701)46:1<11::AID-MRD3>3.0.CO;2-U. Mol Reprod Dev. 1997. PMID: 8981358 Review.
Cited by
-
Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34.J Leukoc Biol. 2014 Dec;96(6):1143-53. doi: 10.1189/jlb.4A0614-295R. Epub 2014 Sep 4. J Leukoc Biol. 2014. PMID: 25190077 Free PMC article.
-
Functions of interleukin-34 and its emerging association with rheumatoid arthritis.Immunology. 2016 Dec;149(4):362-373. doi: 10.1111/imm.12660. Epub 2016 Sep 23. Immunology. 2016. PMID: 27550090 Free PMC article. Review.
-
IL-34-Dependent Intrarenal and Systemic Mechanisms Promote Lupus Nephritis in MRL-Faslpr Mice.J Am Soc Nephrol. 2019 Feb;30(2):244-259. doi: 10.1681/ASN.2018090901. Epub 2019 Jan 8. J Am Soc Nephrol. 2019. PMID: 30622154 Free PMC article.
-
Immunoregulatory properties of the cytokine IL-34.Cell Mol Life Sci. 2017 Jul;74(14):2569-2586. doi: 10.1007/s00018-017-2482-4. Epub 2017 Mar 3. Cell Mol Life Sci. 2017. PMID: 28258292 Free PMC article. Review.
-
Mechanisms of Fish Macrophage Antimicrobial Immunity.Front Immunol. 2018 May 28;9:1105. doi: 10.3389/fimmu.2018.01105. eCollection 2018. Front Immunol. 2018. PMID: 29892285 Free PMC article. Review.
References
-
- Vinuesa C. G., Sanz I., Cook M. C. (2009) Dysregulation of germinal centres in autoimmune disease. Nat. Rev. Immunol. 9, 845–857 - PubMed
-
- Rajewsky K. (1996) Clonal selection and learning in the antibody system. Nature 381, 751–758 - PubMed
-
- Liu M., Schatz D. G. (2009) Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 30, 173–181 - PubMed
-
- Kosco-Vilbois M. H. (2003) Are follicular dendritic cells really good for nothing? Nat. Rev. Immunol. 3, 764–769 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/A/00001660/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/A/00001659/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P30 CA013330/CA/NCI NIH HHS/United States
- BBS/E/D/20251969/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 CA032551/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

