Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells
- PMID: 24052944
- PMCID: PMC3757747
- DOI: 10.1016/j.stemcr.2013.03.005
Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells
Abstract
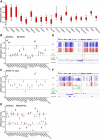
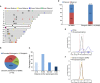
Parental imprinting is an epigenetic phenomenon by which genes are expressed in a monoallelic fashion, according to their parent of origin. DNA methylation is considered the hallmark mechanism regulating parental imprinting. To identify imprinted differentially methylated regions (DMRs), we compared the DNA methylation status between multiple normal and parthenogenetic human pluripotent stem cells (PSCs) by performing reduced representation bisulfite sequencing. Our analysis identified over 20 previously unknown imprinted DMRs in addition to the known DMRs. These include DMRs in loci associated with human disorders, and a class of intergenic DMRs that do not seem to be related to gene expression. Furthermore, the study showed some DMRs to be unstable, liable to differentiation or reprogramming. A comprehensive comparison between mouse and human DMRs identified almost half of the imprinted DMRs to be species specific. Taken together, our data map novel DMRs in the human genome, their evolutionary conservation, and relation to gene expression.
Figures


References
-
- Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
-
- Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

