Myeloproliferative neoplasm with ETV6-ABL1 fusion: a case report and literature review
- PMID: 24053143
- PMCID: PMC3853649
- DOI: 10.1186/1755-8166-6-39
Myeloproliferative neoplasm with ETV6-ABL1 fusion: a case report and literature review
Abstract
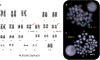
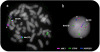
ETV6-ABL1 is a rare gene fusion with oncogenic properties, reported so far in 28 patients presenting a variety of haematological malignancies associated with clinical outcome, including chronic myeloid leukaemia (CML), acute myeloid leukaemia (AML), acute lymphoblastic leukaemia (ALL) and chronic myeloproliferative neoplasm (cMPN). Here we report on a 46-year-old female who presented with Philadelphia negative CML, positive for the ETV6-ABL1 fusion. Whole genome screening carried out with oligonucleotide arrays showed a subtle loss at 12p13 and cryptic imbalances within the 9q34.3 region in a highly unstable genome. FISH mapping with custom BAC probes identified two breakpoints 5 Mb apart within the 9q34 region, together with a break at 12p13. While FISH with commercial BCR-ABL1 probes failed to detect any ABL1 changes, the ETV6 break-apart probe conclusively identified the ETV6-ABL1 fusion thus determining the probe's role as the primary diagnostic FISH test for this chimeric oncogene. In addition, we confirm the association of the ETV6-ABL1 fusion with imatinib resistance reported so far in three other patients, while recording excellent response to the 2nd generation tyrosine kinase inhibitor (TKI) nilotinib. In summary, we highlight the value of ETV6 FISH as a diagnostic test and the therapy resistance of ETV6-ABL1 positive disorders to imatinib.
Figures


References
-
- Papadopoulos P, Ridge SA, Boucher CA, Stocking C, Wiedemann LM. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

