Novel aspects of glutamine synthetase (GS) regulation revealed by a detailed expression analysis of the entire GS gene family of Medicago truncatula under different physiological conditions
- PMID: 24053168
- PMCID: PMC3848809
- DOI: 10.1186/1471-2229-13-137
Novel aspects of glutamine synthetase (GS) regulation revealed by a detailed expression analysis of the entire GS gene family of Medicago truncatula under different physiological conditions
Abstract
Background: Glutamine Synthetase (GS, EC 6.3.1.2) is a central enzyme in nitrogen metabolism, and a key component of nitrogen use efficiency (NUE) and plant yield and thus it is extremely important to understand how it is regulated in plants. Medicago truncatula provides an excellent model system to study GS, as it contain a very simple GS gene family comprising only four expressed genes, MtGS1a and MtGS1b encoding cytosolic polypeptides, and MtGS2a and MtGS2b encoding plastid-located enzymes. To identify new regulatory mechanisms controlling GS activity, we performed a detailed expression analysis of the entire GS gene family of M. truncatula in the major organs of the plant, over a time course of nodule or seed development and during a diurnal cycle.
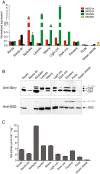
Results: Individual GS transcripts were quantified by qRT-PCR, and GS polypeptides and holoenzymes were evaluated by western blot and in-gel activity under native electrophoresis. These studies revealed that all four GS genes are differentially regulated in each organ of the plant, in a developmental manner, and identified new regulatory controls, which appear to be specific to certain metabolic contexts. Studies of the protein profiles showed that the GS polypeptides assemble into organ-specific protein complexes and suffer organ-specific post-translational modifications under defined physiological conditions. Our studies also reveal that GS expression and activity are modulated during a diurnal cycle. The biochemical properties of the four isoenzymes were determined and are discussed in relation to their function in the plant.
Conclusions: This work provides a comprehensive overview of GS expression and regulation in the model legume M. truncatula, contributing to a better understanding of the specific function of individual isoenzymes and to the identification of novel organ-specific post-translational mechanisms of GS regulation. We demonstrate that the GS proteins are modified and/or integrated into protein-complexes that assemble into a specific composition in particular organs of the plant. Taken together, the results presented here open new avenues to explore the regulatory mechanisms controlling GS activity in plants, a subject of major importance due to the crucial importance of the enzyme for plant growth and productivity.
Figures
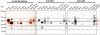




References
-
- Lea PJ, Miflin BJ. Nitrogen assimilation and its relevance to crop improvement. Annu Plant Rev:Nitrogen Metab Plants in the Post-Genom Era. 2010;42:1–40.
-
- Hirel B, Lea P. In: Plant nitrogen. Morot-Gaudry PSL, editor. Berlin: Springer-Verlag; 2001. Ammonia assimilation; pp. 779–799.
-
- Forde BG, Cullimore JV. The molecular biology of glutamine synthetase in higher plants. Oxf surv of plant mol cell biol. 1989;6:49.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

