Extremes of lineage plasticity in the Drosophila brain
- PMID: 24055154
- PMCID: PMC3902790
- DOI: 10.1016/j.cub.2013.07.074
Extremes of lineage plasticity in the Drosophila brain
Abstract
An often-overlooked aspect of neural plasticity is the plasticity of neuronal composition, in which the numbers of neurons of particular classes are altered in response to environment and experience. The Drosophila brain features several well-characterized lineages in which a single neuroblast gives rise to multiple neuronal classes in a stereotyped sequence during development. We find that in the intrinsic mushroom body neuron lineage, the numbers for each class are highly plastic, depending on the timing of temporal fate transitions and the rate of neuroblast proliferation. For example, mushroom body neuroblast cycling can continue under starvation conditions, uncoupled from temporal fate transitions that depend on extrinsic cues reflecting organismal growth and development. In contrast, the proliferation rates of antennal lobe lineages are closely associated with organismal development, and their temporal fate changes appear to be cell cycle-dependent, such that the same numbers and types of uniglomerular projection neurons innervate the antennal lobe following various perturbations. We propose that this surprising difference in plasticity for these brain lineages is adaptive, given their respective roles as parallel processors versus discrete carriers of olfactory information.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
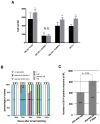
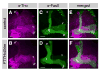
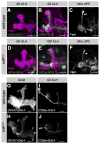
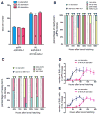
Similar articles
-
Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system.Dev Biol. 2013 Jan 15;373(2):322-37. doi: 10.1016/j.ydbio.2012.11.003. Epub 2012 Nov 10. Dev Biol. 2013. PMID: 23149077 Free PMC article.
-
The zinc finger transcription factor Jing is required for dendrite/axonal targeting in Drosophila antennal lobe development.Dev Biol. 2013 Sep 1;381(1):17-27. doi: 10.1016/j.ydbio.2013.06.023. Epub 2013 Jun 25. Dev Biol. 2013. PMID: 23810656
-
Metamorphosis of an identified serotonergic neuron in the Drosophila olfactory system.Neural Dev. 2007 Oct 24;2:20. doi: 10.1186/1749-8104-2-20. Neural Dev. 2007. PMID: 17958902 Free PMC article.
-
Brn3/POU-IV-type POU homeobox genes-Paradigmatic regulators of neuronal identity across phylogeny.Wiley Interdiscip Rev Dev Biol. 2020 Jul;9(4):e374. doi: 10.1002/wdev.374. Epub 2020 Feb 3. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32012462 Review.
-
The role of the Drosophila lateral horn in olfactory information processing and behavioral response.J Insect Physiol. 2017 Apr;98:29-37. doi: 10.1016/j.jinsphys.2016.11.007. Epub 2016 Nov 19. J Insect Physiol. 2017. PMID: 27871975 Review.
Cited by
-
Nucleolar stress in Drosophila neuroblasts, a model for human ribosomopathies.Biol Open. 2020 Apr 13;9(4):bio046565. doi: 10.1242/bio.046565. Biol Open. 2020. PMID: 32184230 Free PMC article.
-
Environmental effects on Drosophila brain development and learning.J Exp Biol. 2018 Jan 10;221(Pt 1):jeb169375. doi: 10.1242/jeb.169375. J Exp Biol. 2018. PMID: 29061687 Free PMC article.
-
Maintaining robust size across environmental conditions through plastic brain growth dynamics.Open Biol. 2022 Sep;12(9):220037. doi: 10.1098/rsob.220037. Epub 2022 Sep 14. Open Biol. 2022. PMID: 36102061 Free PMC article.
-
Roles of Hox genes in the patterning of the central nervous system of Drosophila.Fly (Austin). 2014;8(1):26-32. doi: 10.4161/fly.27424. Epub 2013 Dec 5. Fly (Austin). 2014. PMID: 24406332 Free PMC article.
-
Eyeless uncouples mushroom body neuroblast proliferation from dietary amino acids in Drosophila.Elife. 2017 Aug 9;6:e26343. doi: 10.7554/eLife.26343. Elife. 2017. PMID: 28826476 Free PMC article.
References
-
- Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003:266–275. - PubMed
-
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;18:4065–4076. - PubMed
-
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;1:145–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

