Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S
- PMID: 24055344
- PMCID: PMC3825818
- DOI: 10.1016/j.molcel.2013.08.024
Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S
Abstract
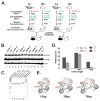
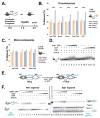
Chromatin remodelers have been implicated in the regulation of histone-modifying complexes. However, the underlying mechanism remains poorly understood. The Rpd3S histone deacetylase complex is recruited by elongating RNA polymerase II to remove histone acetylation at coding regions in a manner that is dependent on methylation of lysine 36 on histone 3 (H3K36me), and Rpd3S prefers dinucleosomes. Here, we show that the binding of Rpd3S to dinucleosomes and its catalytic activity are sensitive to the length of nucleosomal linker in a nonlinear fashion. Intriguingly, we found that H3K36me on one nucleosome stimulates Rpd3S to deacetylate the neighboring nucleosomes when those two nucleosomes are within an optimal distance. Finally, we demonstrate that chromatin remodelers enhance Rpd3S activity by altering nucleosomal spacing, suggesting that chromatin remodelers prime chromatin configuration to fine-tune subsequent histone modification reactions. This mechanism is important for accurate temporal control of chromatin dynamics during the transcription elongation cycle.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


References
-
- Carre C, Ciurciu A, Komonyi O, Jacquier C, Fagegaltier D, Pidoux J, Tricoire H, Tora L, Boros IM, Antoniewski C. The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO reports. 2008;9:187–192. - PMC - PubMed
-
- Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005;1731:77–87. discussion 75–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

