Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus
- PMID: 24056699
- PMCID: PMC4974078
- DOI: 10.1038/nn.3522
Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus
Abstract
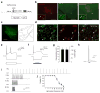
Rapid-eye movement (REM) sleep correlates with neuronal activity in the brainstem, basal forebrain and lateral hypothalamus. Lateral hypothalamus melanin-concentrating hormone (MCH)-expressing neurons are active during sleep, but their effects on REM sleep remain unclear. Using optogenetic tools in newly generated Tg(Pmch-cre) mice, we found that acute activation of MCH neurons (ChETA, SSFO) at the onset of REM sleep extended the duration of REM, but not non-REM, sleep episodes. In contrast, their acute silencing (eNpHR3.0, archaerhodopsin) reduced the frequency and amplitude of hippocampal theta rhythm without affecting REM sleep duration. In vitro activation of MCH neuron terminals induced GABAA-mediated inhibitory postsynaptic currents in wake-promoting histaminergic neurons of the tuberomammillary nucleus (TMN), and in vivo activation of MCH neuron terminals in TMN or medial septum also prolonged REM sleep episodes. Collectively, these results suggest that activation of MCH neurons maintains REM sleep, possibly through inhibition of arousal circuits in the mammalian brain.
Figures
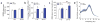




References
-
- JOUVET M. Research on the neural structures and responsible mechanisms in different phases of physiological sleep. Arch Ital Biol. 1962;100:125–206. - PubMed
-
- McGinty DJ, Harper RM. Dorsal raphe neurons: depression of firing during sleep in cats. Brain Res. 1976;101:569–575. - PubMed
-
- McCarley RW, Hobson JA. Neuronal excitability modulation over the sleep cycle: a structural and mathematical model. Science. 1975;189:58–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

