Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation
- PMID: 24056746
- PMCID: PMC3805781
- DOI: 10.1038/ni.2712
Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation
Abstract
Although intergenic long noncoding RNAs (lincRNAs) have been linked to gene regulation in various tissues, little is known about lincRNA transcriptomes in the T cell lineages. Here we identified 1,524 lincRNA clusters in 42 T cell samples, from early T cell progenitors to terminally differentiated helper T cell subsets. Our analysis revealed highly dynamic and cell-specific expression patterns for lincRNAs during T cell differentiation. These lincRNAs were located in genomic regions enriched for genes that encode proteins with immunoregulatory functions. Many were bound and regulated by the key transcription factors T-bet, GATA-3, STAT4 and STAT6. We found that the lincRNA LincR-Ccr2-5'AS, together with GATA-3, was an essential component of a regulatory circuit in gene expression specific to the TH2 subset of helper T cells and was important for the migration of TH2 cells.
Figures
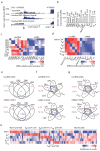
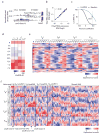


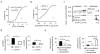

References
-
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–72. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

