Integrated conformational and lipid-sensing regulation of endosomal ArfGEF BRAG2
- PMID: 24058294
- PMCID: PMC3769224
- DOI: 10.1371/journal.pbio.1001652
Integrated conformational and lipid-sensing regulation of endosomal ArfGEF BRAG2
Abstract
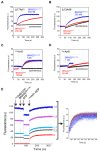
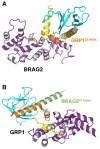
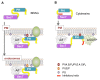
The mechanisms whereby guanine nucleotide exchange factors (GEFs) coordinate their subcellular targeting to their activation of small GTPases remain poorly understood. Here we analyzed how membranes control the efficiency of human BRAG2, an ArfGEF involved in receptor endocytosis, Wnt signaling, and tumor invasion. The crystal structure of an Arf1-BRAG2 complex that mimics a membrane-bound intermediate revealed an atypical PH domain that is constitutively anchored to the catalytic Sec7 domain and interacts with Arf. Combined with the quantitative analysis of BRAG2 exchange activity reconstituted on membranes, we find that this PH domain potentiates nucleotide exchange by about 2,000-fold by cumulative conformational and membrane-targeting contributions. Furthermore, it restricts BRAG2 activity to negatively charged membranes without phosphoinositide specificity, using a positively charged surface peripheral to but excluding the canonical lipid-binding pocket. This suggests a model of BRAG2 regulation along the early endosomal pathway that expands the repertoire of GEF regulatory mechanisms. Notably, it departs from the auto-inhibitory and feedback loop paradigm emerging from studies of SOS and cytohesins. It also uncovers a novel mechanism of unspecific lipid-sensing by PH domains that may allow sustained binding to maturating membranes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Expanding the regulatory repertoire of GEFs.PLoS Biol. 2013 Sep;11(9):e1001654. doi: 10.1371/journal.pbio.1001654. Epub 2013 Sep 10. PLoS Biol. 2013. PMID: 24058296 Free PMC article. No abstract available.
References
-
- D'Souza-Schorey C, Chavrier P (2006) ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7: 347–358. - PubMed
-
- Cherfils J, Zeghouf M (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309. - PubMed
-
- Renault L, Guibert B, Cherfils J (2003) Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426: 525–530. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

