SNP discovery by illumina-based transcriptome sequencing of the olive and the genetic characterization of Turkish olive genotypes revealed by AFLP, SSR and SNP markers
- PMID: 24058483
- PMCID: PMC3772808
- DOI: 10.1371/journal.pone.0073674
SNP discovery by illumina-based transcriptome sequencing of the olive and the genetic characterization of Turkish olive genotypes revealed by AFLP, SSR and SNP markers
Abstract
Background: The olive tree (Olea europaea L.) is a diploid (2n = 2x = 46) outcrossing species mainly grown in the Mediterranean area, where it is the most important oil-producing crop. Because of its economic, cultural and ecological importance, various DNA markers have been used in the olive to characterize and elucidate homonyms, synonyms and unknown accessions. However, a comprehensive characterization and a full sequence of its transcriptome are unavailable, leading to the importance of an efficient large-scale single nucleotide polymorphism (SNP) discovery in olive. The objectives of this study were (1) to discover olive SNPs using next-generation sequencing and to identify SNP primers for cultivar identification and (2) to characterize 96 olive genotypes originating from different regions of Turkey.
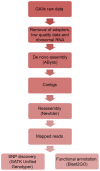
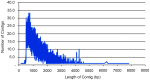
Methodology/principal findings: Next-generation sequencing technology was used with five distinct olive genotypes and generated cDNA, producing 126,542,413 reads using an Illumina Genome Analyzer IIx. Following quality and size trimming, the high-quality reads were assembled into 22,052 contigs with an average length of 1,321 bases and 45 singletons. The SNPs were filtered and 2,987 high-quality putative SNP primers were identified. The assembled sequences and singletons were subjected to BLAST similarity searches and annotated with a Gene Ontology identifier. To identify the 96 olive genotypes, these SNP primers were applied to the genotypes in combination with amplified fragment length polymorphism (AFLP) and simple sequence repeats (SSR) markers.
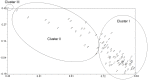
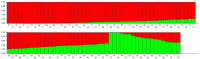
Conclusions/significance: This study marks the highest number of SNP markers discovered to date from olive genotypes using transcriptome sequencing. The developed SNP markers will provide a useful source for molecular genetic studies, such as genetic diversity and characterization, high density quantitative trait locus (QTL) analysis, association mapping and map-based gene cloning in the olive. High levels of genetic variation among Turkish olive genotypes revealed by SNPs, AFLPs and SSRs allowed us to characterize the Turkish olive genotype.
Conflict of interest statement
Figures




References
-
- Zohary D, Hopf M (2000) Domestication of plants in the old world: The origin and spread of cultivated plants in West Asia, Europe, and the Nile Valley Oxford University Press, NY.
-
- Green PS (2002) A revision of Olea L. (Oleaceae). Kew Bull 57.
-
- Bartolini L, Prevost G, Messeri C, Carignani G, Menini U (1998) Olive germplasm: Cultivars and worldwide collections. Food and Agriculture Organization.
-
- Boskou G, Salta FN, Chrysostomou S, Mylona A, Chiou A, et al... (2006) Antioxidant capacity and phenolic profile of table olives from the Greek market. Food Chemistry 94 558–564.
-
- Visioli F, Galli C (2002) Biological properties of olive oil phytochemicals. Critical Reviews of Food Science and Nutrition. 42 3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

