Hepatitis C virus replication and Golgi function in brefeldin a-resistant hepatoma-derived cells
- PMID: 24058576
- PMCID: PMC3776844
- DOI: 10.1371/journal.pone.0074491
Hepatitis C virus replication and Golgi function in brefeldin a-resistant hepatoma-derived cells
Abstract
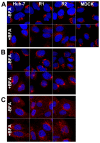
Recent reports indicate that the replication of hepatitis C virus (HCV) depends on the GBF1-Arf1-COP-I pathway. We generated Huh-7-derived cell lines resistant to brefeldin A (BFA), which is an inhibitor of this pathway. The resistant cell lines could be sorted into two phenotypes regarding BFA-induced toxicity, inhibition of albumin secretion, and inhibition of HCV infection. Two cell lines were more than 100 times more resistant to BFA than the parental Huh-7 cells in these 3 assays. This resistant phenotype was correlated with the presence of a point mutation in the Sec7 domain of GBF1, which is known to impair the binding of BFA. Surprisingly, the morphology of the cis-Golgi of these cells remained sensitive to BFA at concentrations of the drug that allowed albumin secretion, indicating a dichotomy between the phenotypes of secretion and Golgi morphology. Cells of the second group were about 10 times more resistant than parental Huh-7 cells to the BFA-induced toxicity. The EC50 for albumin secretion was only 1.5-1.8 fold higher in these cells than in Huh-7 cells. However their level of secretion in the presence of inhibitory doses of BFA was 5 to 15 times higher. Despite this partially effective secretory pathway in the presence of BFA, the HCV infection was almost as sensitive to BFA as in Huh-7 cells. This suggests that the function of GBF1 in HCV replication does not simply reflect its role of regulator of the secretory pathway of the host cell. Thus, our results confirm the involvement of GBF1 in HCV replication, and suggest that GBF1 might fulfill another function, in addition to the regulation of the secretory pathway, during HCV replication.
Conflict of interest statement
Figures








References
-
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, et al. (2003) Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77: 5487–5492 doi:10.1128/JVI.77.9.5487-5492.2003 - DOI - PMC - PubMed
-
- Rouillé Y, Helle F, Delgrange D, Roingeard P, Voisset C, et al. (2006) Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol 80: 2832–2841 doi:10.1128/JVI.80.6.2832-2841.2006 - DOI - PMC - PubMed
-
- Wölk B, Büchele B, Moradpour D, Rice CM (2008) A dynamic view of hepatitis C virus replication complexes. J Virol 82: 10519–10531 doi:10.1128/JVI.00640-08 - DOI - PMC - PubMed
-
- Ferraris P, Blanchard E, Roingeard P (2010) Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. Journal of General Virology 91: 2230–2237 doi:10.1099/vir.0.022186-0 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Medical
