Sequence recombination and conservation of Varroa destructor virus-1 and deformed wing virus in field collected honey bees (Apis mellifera)
- PMID: 24058580
- PMCID: PMC3776811
- DOI: 10.1371/journal.pone.0074508
Sequence recombination and conservation of Varroa destructor virus-1 and deformed wing virus in field collected honey bees (Apis mellifera)
Abstract
We sequenced small (s) RNAs from field collected honeybees (Apis mellifera) and bumblebees (Bombuspascuorum) using the Illumina technology. The sRNA reads were assembled and resulting contigs were used to search for virus homologues in GenBank. Matches with Varroadestructor virus-1 (VDV1) and Deformed wing virus (DWV) genomic sequences were obtained for A. mellifera but not B. pascuorum. Further analyses suggested that the prevalent virus population was composed of VDV-1 and a chimera of 5'-DWV-VDV1-DWV-3'. The recombination junctions in the chimera genomes were confirmed by using RT-PCR, cDNA cloning and Sanger sequencing. We then focused on conserved short fragments (CSF, size > 25 nt) in the virus genomes by using GenBank sequences and the deep sequencing data obtained in this study. The majority of CSF sites confirmed conservation at both between-species (GenBank sequences) and within-population (dataset of this study) levels. However, conserved nucleotide positions in the GenBank sequences might be variable at the within-population level. High mutation rates (Pi>10%) were observed at a number of sites using the deep sequencing data, suggesting that sequence conservation might not always be maintained at the population level. Virus-host interactions and strategies for developing RNAi treatments against VDV1/DWV infections are discussed.
Conflict of interest statement
Figures

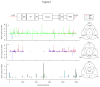
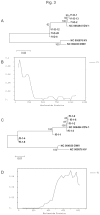

References
-
- Genersch E, Aubert M (2010) Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Vet Res 41: 54. doi:10.1051/vetres/2010027. PubMed: 20423694. - DOI - PMC - PubMed
-
- Evans JD, Schwarz RS (2011) Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol 19: 614-620. doi:10.1016/j.tim.2011.09.003. PubMed: 22032828. - DOI - PubMed
-
- Yang X, Cox-Foster D (2007) Effects of parasitization by Varroa destructor on survivorship and physiological traits of Apis mellifera in correlation with viral incidence and microbial challenge. Parasitology 134: 405-412. doi:10.1017/S0031182006000710. PubMed: 17078903. - DOI - PubMed
-
- Yang X, Cox-Foster DL (2005) Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc Natl Acad Sci U S A 102: 7470-7475. doi:10.1073/pnas.0501860102. PubMed: 15897457. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

