Genetic diversity in black South Africans from Soweto
- PMID: 24059264
- PMCID: PMC3850641
- DOI: 10.1186/1471-2164-14-644
Genetic diversity in black South Africans from Soweto
Abstract
Background: Due to the unparalleled genetic diversity of its peoples, Africa is attracting growing research attention. Several African populations have been assessed in global initiatives such as the International HapMap and 1000 Genomes Projects. Notably excluded, however, is the southern Africa region, which is inhabited predominantly by southeastern Bantu-speakers, currently suffering under the dual burden of infectious and non-communicable diseases. Limited reference data for these individuals hampers medical research and prevents thorough understanding of the underlying population substructure. Here, we present the most detailed exploration, to date, of genetic diversity in 94 unrelated southeastern Bantu-speaking South Africans, resident in urban Soweto (Johannesburg).
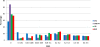
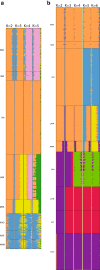
Results: Participants were typed for ~4.3 million SNPs using the Illumina Omni5 beadchip. PCA and ADMIXTURE plots were used to compare the observed variation with that seen in selected populations worldwide. Results indicated that Sowetans, and other southeastern Bantu-speakers, are a clearly distinct group from other African populations previously investigated, reflecting a unique genetic history with small, but significant contributions from diverse sources. To assess the suitability of our sample as representative of Sowetans, we compared our results to participants in a larger rheumatoid arthritis case-control study. The control group showed good clustering with our sample, but among the cases were individuals who demonstrated notable admixture.
Conclusions: Sowetan population structure appears unique compared to other black Africans, and may have clinical implications. Our data represent a suitable reference set for southeastern Bantu-speakers, on par with a HapMap type reference population, and constitute a prelude to the Southern African Human Genome Programme.
Figures



References
-
- Cann HM, de-Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A. et al.A human genome diversity cell line panel. Science. 2002;296:261–262. - PubMed
-
- Pagani L, Kivisild T, Tarekegn A, Ekong R, Plaster C, Gallego Romero I, Ayub Q, Mehdi SQ, Thomas MG, Luiselli D. et al.Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

