KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors
- PMID: 24059286
- PMCID: PMC3766012
- DOI: 10.1186/1471-2334-13-405
KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors
Abstract
Background: Natural killer (NK) cells have emerged as pivotal players in innate immunity, especially in the defense against viral infections and tumors. Killer immunoglobulin-like receptors (KIRs)--an important recognition receptor expressed on the surface of NK cells--regulate the inhibition and/or activation of NK cells after interacting with human leukocyte antigen (HLA) class I ligands. Various KIR genes might impact the prognosis of many different diseases. The implications of KIR-HLA interaction in HIV disease progression remains poorly understood.
Methods: Here, we studied KIR genotypes, mRNA levels, HLA genotypes, CD4+ T cell counts and viral loads in our cohort of Human Immunodeficiency Virus (HIV)-infected individuals, a group that includes HIV long-term nonprogressors (LTNPs) and typical progressors (TPs).
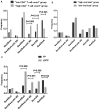
Results: We found that the frequency of KIR3DS1/L1 heterozygotes with HLA-Bw4-80I gene was much higher in LTNPs than in TPs (P = 0.001) and that the KIR3DL1 homozygotes without HLA-Bw4-80I gene had higher viral loads and lower CD4+ T cell counts (P = 0.014 and P = 0.021, respectively). Our study also confirmed that homozygosity for the HLA-Bw6 allele was associated with rapid disease progression. In addition to the aforementioned results on the DNA level, we observed that higher level expression of KIR3DS1 mRNA was in LTNP group, and that higher level expression of KIR3DL1 mRNA was in TP group.
Conclusions: Our data suggest that different KIR-HLA genotypes and different levels of transcripts associate with HIV disease progression.
Figures

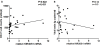
Similar articles
-
Protective genotypes in HIV infection reflect superior function of KIR3DS1+ over KIR3DL1+ CD8+ T cells.Immunol Cell Biol. 2015 Jan;93(1):67-76. doi: 10.1038/icb.2014.68. Epub 2014 Aug 12. Immunol Cell Biol. 2015. PMID: 25112829 Free PMC article.
-
Inhibitory natural killer cell receptor KIR3DL1 with its ligand Bw4 constraints HIV-1 disease among South Indians.AIDS. 2018 Nov 28;32(18):2679-2688. doi: 10.1097/QAD.0000000000002028. AIDS. 2018. PMID: 30289808
-
Time to seroconversion in HIV-exposed subjects carrying protective versus non protective KIR3DS1/L1 and HLA-B genotypes.PLoS One. 2014 Oct 17;9(10):e110480. doi: 10.1371/journal.pone.0110480. eCollection 2014. PLoS One. 2014. PMID: 25330014 Free PMC article.
-
KIR-HLA intercourse in HIV disease.Trends Microbiol. 2008 Dec;16(12):620-7. doi: 10.1016/j.tim.2008.09.002. Epub 2008 Oct 29. Trends Microbiol. 2008. PMID: 18976921 Free PMC article. Review.
-
The role genes encoding of killer cell immunoglobulin-like receptors (KIRs) and their ligands in susceptibility to and progression of HIV infection.Postepy Hig Med Dosw (Online). 2016 Dec 31;70(0):1409-1423. doi: 10.5604/17322693.1227771. Postepy Hig Med Dosw (Online). 2016. PMID: 28100849 Review.
Cited by
-
HLA-F on Autologous HIV-Infected Cells Activates Primary NK Cells Expressing the Activating Killer Immunoglobulin-Like Receptor KIR3DS1.J Virol. 2019 Aug 28;93(18):e00933-19. doi: 10.1128/JVI.00933-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31270222 Free PMC article.
-
High-resolution KIR and HLA genotyping in three Chinese ethnic minorities reveals distinct origins.HLA. 2024 Apr;103(4):e15482. doi: 10.1111/tan.15482. HLA. 2024. PMID: 38625090 Free PMC article.
-
A copy number variation map of the human genome.Nat Rev Genet. 2015 Mar;16(3):172-83. doi: 10.1038/nrg3871. Epub 2015 Feb 3. Nat Rev Genet. 2015. PMID: 25645873 Review.
-
Unmapped exome reads implicate a role for Anelloviridae in childhood HIV-1 long-term non-progression.NPJ Genom Med. 2021 Mar 19;6(1):24. doi: 10.1038/s41525-021-00185-w. NPJ Genom Med. 2021. PMID: 33741997 Free PMC article.
-
Regulation and Function of NK and T Cells During Dengue Virus Infection and Vaccination.Adv Exp Med Biol. 2018;1062:251-264. doi: 10.1007/978-981-10-8727-1_18. Adv Exp Med Biol. 2018. PMID: 29845538 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

