Positive and negative phosphorylation regulates RIP1- and RIP3-induced programmed necrosis
- PMID: 24059293
- PMCID: PMC4143978
- DOI: 10.1042/BJ20130860
Positive and negative phosphorylation regulates RIP1- and RIP3-induced programmed necrosis
Abstract
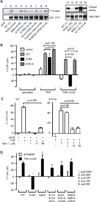
Programmed necrosis or necroptosis is controlled by the action of two serine/threonine kinases, RIP1 (receptor-interacting serine/threonine protein kinase 1; also known as RIPK1) and RIP3. The phosphorylation of RIP1 and RIP3 is critical for assembly of the necrosome, an amyloid-like complex that initiates transmission of the pro-necrotic signal. In the present study, we used site-directed mutagenesis to systematically examine the effects of putative phosphoacceptor sites on RIP1 and RIP3 on TNF (tumour necrosis factor)-induced programmed necrosis. We found that mutation of individual serine residues in the kinase domain of RIP1 had little effect on RIP1 kinase activity and TNF-induced programmed necrosis. Surprisingly, an alanine residue substitution for Ser(89) enhanced RIP1 kinase activity and TNF-induced programmed necrosis without affecting RIP1-RIP3 necrosome formation. This indicates that Ser(89) is an inhibitory phosphoacceptor site that can dampen the pro-necrotic function of RIP1. In addition, we show that a phosphomimetic mutant of RIP3, S204D, led to programmed necrosis that was refractory to RIP1 siRNA and insensitive to necrostatin-1 inhibition. Our results show that programmed necrosis is regulated by positive and inhibitory phosphorylation events.
Figures

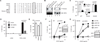
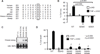
References
-
- Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. - PubMed
-
- Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

