Evolution of the myosin heavy chain gene MYH14 and its intronic microRNA miR-499: muscle-specific miR-499 expression persists in the absence of the ancestral host gene
- PMID: 24059862
- PMCID: PMC3716903
- DOI: 10.1186/1471-2148-13-142
Evolution of the myosin heavy chain gene MYH14 and its intronic microRNA miR-499: muscle-specific miR-499 expression persists in the absence of the ancestral host gene
Abstract
Background: A novel sarcomeric myosin heavy chain gene, MYH14, was identified following the completion of the human genome project. MYH14 contains an intronic microRNA, miR-499, which is expressed in a slow/cardiac muscle specific manner along with its host gene; it plays a key role in muscle fiber-type specification in mammals. Interestingly, teleost fish genomes contain multiple MYH14 and miR-499 paralogs. However, the evolutionary history of MYH14 and miR-499 has not been studied in detail. In the present study, we identified MYH14/miR-499 loci on various teleost fish genomes and examined their evolutionary history by sequence and expression analyses.
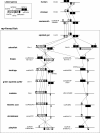
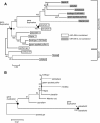
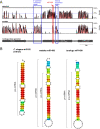
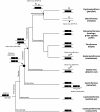
Results: Synteny and phylogenetic analyses depict the evolutionary history of MYH14/miR-499 loci where teleost specific duplication and several subsequent rounds of species-specific gene loss events took place. Interestingly, miR-499 was not located in the MYH14 introns of certain teleost fish. An MYH14 paralog, lacking miR-499, exhibited an accelerated rate of evolution compared with those containing miR-499, suggesting a putative functional relationship between MYH14 and miR-499. In medaka, Oryzias latipes, miR-499 is present where MYH14 is completely absent in the genome. Furthermore, by using in situ hybridization and small RNA sequencing, miR-499 was expressed in the notochord at the medaka embryonic stage and slow/cardiac muscle at the larval and adult stages. Comparing the flanking sequences of MYH14/miR-499 loci between torafugu Takifugu rubripes, zebrafish Danio rerio, and medaka revealed some highly conserved regions, suggesting that cis-regulatory elements have been functionally conserved in medaka miR-499 despite the loss of its host gene.
Conclusions: This study reveals the evolutionary history of the MYH14/miRNA-499 locus in teleost fish, indicating divergent distribution and expression of MYH14 and miR-499 genes in different teleost fish lineages. We also found that medaka miR-499 was even expressed in the absence of its host gene. To our knowledge, this is the first report that shows the conversion of intronic into non-intronic miRNA during the evolution of a teleost fish lineage.
Figures

References
-
- Shrager JB, Desjardins PR, Burkman JM, Konig SK, Stewart SK, Su L, Shah MC, Bricklin E, Tewari M, Hoffman R, Rickels MR, Jullian EH, Rubinstein NA, Stedman HH. Human skeletal myosin heavy chain genes are tightly linked in the order embryonic-IIa-IId/x-ILb-perinatal-extraocular. J Muscle Res Cell Motil. 2000;21:345–355. doi: 10.1023/A:1005635030494. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

