Sharing the slope: depth partitioning of agariciid corals and associated Symbiodinium across shallow and mesophotic habitats (2-60 m) on a Caribbean reef
- PMID: 24059868
- PMCID: PMC3849765
- DOI: 10.1186/1471-2148-13-205
Sharing the slope: depth partitioning of agariciid corals and associated Symbiodinium across shallow and mesophotic habitats (2-60 m) on a Caribbean reef
Abstract
Background: Scleractinian corals and their algal endosymbionts (genus Symbiodinium) exhibit distinct bathymetric distributions on coral reefs. Yet, few studies have assessed the evolutionary context of these ecological distributions by exploring the genetic diversity of closely related coral species and their associated Symbiodinium over large depth ranges. Here we assess the distribution and genetic diversity of five agariciid coral species (Agaricia humilis, A. agaricites, A. lamarcki, A. grahamae, and Helioseris cucullata) and their algal endosymbionts (Symbiodinium) across a large depth gradient (2-60 m) covering shallow to mesophotic depths on a Caribbean reef.
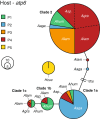
Results: The five agariciid species exhibited distinct depth distributions, and dominant Symbiodinium associations were found to be species-specific, with each of the agariciid species harbouring a distinct ITS2-DGGE profile (except for a shared profile between A. lamarcki and A. grahamae). Only A. lamarcki harboured different Symbiodinium types across its depth distribution (i.e. exhibited symbiont zonation). Phylogenetic analysis (atp6) of the coral hosts demonstrated a division of the Agaricia genus into two major lineages that correspond to their bathymetric distribution ("shallow": A. humilis / A. agaricites and "deep": A. lamarcki / A. grahamae), highlighting the role of depth-related factors in the diversification of these congeneric agariciid species. The divergence between "shallow" and "deep" host species was reflected in the relatedness of the associated Symbiodinium (with A. lamarcki and A. grahamae sharing an identical Symbiodinium profile, and A. humilis and A. agaricites harbouring a related ITS2 sequence in their Symbiodinium profiles), corroborating the notion that brooding corals and their Symbiodinium are engaged in coevolutionary processes.
Conclusions: Our findings support the hypothesis that the depth-related environmental gradient on reefs has played an important role in the diversification of the genus Agaricia and their associated Symbiodinium, resulting in a genetic segregation between coral host-symbiont communities at shallow and mesophotic depths.
Figures






References
-
- Knowlton N, Jackson JBC. New taxonomy and niche partitioning on coral reefs: jack of all trades or master of some? Trends Ecol Evol. 1994;1:7–9. - PubMed
-
- Souter P. Hidden genetic diversity in a key model species of coral. Mar Biol. 2010;157:875–885. doi: 10.1007/s00227-009-1370-3. - DOI
-
- Stefani F, Benzoni F, Yang S-Y, Pichon M, Galli P, Chen CA. Comparison of morphological and genetic analyses reveal cryptic divergence and morphological plasticity in Stylophora (Cnidaria, Scleractinia) Coral Reefs. 2011;30:1033–1049. doi: 10.1007/s00338-011-0797-4. - DOI
-
- Flot J-F, Blanchot J, Charpy L, Cruaud C, Licuanan WY, Nakano Y, Payri C, Tillier S. Incongruence between morphotypes and genetically delimited species in the coral genus Stylophora: phenotypic plasticity, morphological convergence, morphological stasis or interspecific hybridization? BMC Ecol. 2011;11:22. doi: 10.1186/1472-6785-11-22. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

