Genomic distribution of AFLP markers relative to gene locations for different eukaryotic species
- PMID: 24060007
- PMCID: PMC3750350
- DOI: 10.1186/1471-2164-14-528
Genomic distribution of AFLP markers relative to gene locations for different eukaryotic species
Abstract
Background: Amplified fragment length polymorphism (AFLP) markers are frequently used for a wide range of studies, such as genome-wide mapping, population genetic diversity estimation, hybridization and introgression studies, phylogenetic analyses, and detection of signatures of selection. An important issue to be addressed for some of these fields is the distribution of the markers across the genome, particularly in relation to gene sequences.
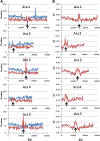
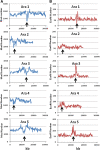
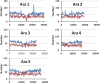
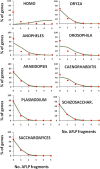
Results: Using in-silico restriction fragment analysis of the genomes of nine eukaryotic species we characterise the distribution of AFLP fragments across the genome and, particularly, in relation to gene locations. First, we identify the physical position of markers across the chromosomes of all species. An observed accumulation of fragments around (peri) centromeric regions in some species is produced by repeated sequences, and this accumulation disappears when AFLP bands rather than fragments are considered. Second, we calculate the percentage of AFLP markers positioned within gene sequences. For the typical EcoRI/MseI enzyme pair, this ranges between 28 and 87% and is usually larger than that expected by chance because of the higher GC content of gene sequences relative to intergenic ones. In agreement with this, the use of enzyme pairs with GC-rich restriction sites substantially increases the above percentages. For example, using the enzyme system SacI/HpaII, 86% of AFLP markers are located within gene sequences in A. thaliana, and 100% of markers in Plasmodium falciparun. We further find that for a typical trait controlled by 50 genes of average size, if 1000 AFLPs are used in a study, the number of those within 1 kb distance from any of the genes would be only about 1-2, and only about 50% of the genes would have markers within that distance.
Conclusions: The high coverage of AFLP markers across the genomes and the high proportion of markers within or close to gene sequences make them suitable for genome scans and detecting large islands of differentiation in the genome. However, for specific traits, the percentage of AFLP markers close to genes can be rather small. Therefore, genome scans directed towards the search of markers closely linked to selected loci can be a difficult task in many instances.
Figures
Similar articles
-
A high-density linkage map in Brassica juncea (Indian mustard) using AFLP and RFLP markers.Theor Appl Genet. 2003 Feb;106(4):607-14. doi: 10.1007/s00122-002-1083-1. Epub 2002 Sep 13. Theor Appl Genet. 2003. PMID: 12595988
-
ATG-anchored AFLP (ATG-AFLP) analysis in cotton.Plant Cell Rep. 2008 Oct;27(10):1645-53. doi: 10.1007/s00299-008-0568-z. Epub 2008 Jun 27. Plant Cell Rep. 2008. PMID: 18584177
-
Beyond an AFLP genome scan towards the identification of immune genes involved in plague resistance in Rattus rattus from Madagascar.Mol Ecol. 2013 Jan;22(2):354-67. doi: 10.1111/mec.12115. Epub 2012 Dec 12. Mol Ecol. 2013. PMID: 23237097
-
Level of polymorphism and genetic mapping of AFLP markers in rice.Genome. 1996 Oct;39(5):969-77. doi: 10.1139/g96-121. Genome. 1996. PMID: 8890522
-
Amplified fragment length polymorphism: an adept technique for genome mapping, genetic differentiation, and intraspecific variation in protozoan parasites.Parasitol Res. 2013 Feb;112(2):457-66. doi: 10.1007/s00436-012-3238-6. Epub 2012 Dec 20. Parasitol Res. 2013. PMID: 23254590 Review.
Cited by
-
Phenotypic responses to light, water, and nutrient conditions in the allopolyploid Arabidopsis suecica and its parent species A. thaliana and A. arenosa: Does the allopolyploid outrange its parents?Ecol Evol. 2022 May 13;12(5):e8915. doi: 10.1002/ece3.8915. eCollection 2022 May. Ecol Evol. 2022. PMID: 35592071 Free PMC article.
-
Fine-scale population epigenetic structure in relation to gastrointestinal parasite load in red grouse (Lagopus lagopus scotica).Mol Ecol. 2014 Sep;23(17):4256-73. doi: 10.1111/mec.12833. Epub 2014 Jul 24. Mol Ecol. 2014. PMID: 24943398 Free PMC article.
-
Genetic Diversity and Population Structure of Endangered Orchid Cypripedium flavum in Fragmented Habitat Using Fluorescent AFLP Markers.Plants (Basel). 2024 Oct 11;13(20):2851. doi: 10.3390/plants13202851. Plants (Basel). 2024. PMID: 39458798 Free PMC article.
References
-
- Bonin A, Ehrich D, Manel S. Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol. 2007;16(18):3737–3758. - PubMed
-
- Meudt HM, Clarke AC. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007;12(3):106–117. - PubMed
-
- Bensch S, Akesson M. Ten years of AFLP in ecology and evolution: why so few animals? Mol Ecol. 2005;14(10):2899–2914. - PubMed
-
- Peters JL, Cnops G, Neyt P, Zethof J, Cornelis K, Van Lijsebettens M, Gerats T. An AFLP-based genome-wide mapping strategy. Theor Appl Genet. 2004;108(2):321–327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

