Optofluidic cell selection from complex microbial communities for single-genome analysis
- PMID: 24060116
- PMCID: PMC3886641
- DOI: 10.1016/B978-0-12-407863-5.00004-6
Optofluidic cell selection from complex microbial communities for single-genome analysis
Abstract
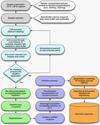
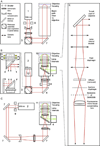
Genetic analysis of single cells is emerging as a powerful approach for studies of heterogeneous cell populations. Indeed, the notion of homogeneous cell populations is receding as approaches to resolve genetic and phenotypic variation between single cells are applied throughout the life sciences. A key step in single-cell genomic analysis today is the physical isolation of individual cells from heterogeneous populations, particularly microbial populations, which often exhibit high diversity. Here, we detail the construction and use of instrumentation for optical trapping inside microfluidic devices to select individual cells for analysis by methods including nucleic acid sequencing. This approach has unique advantages for analyses of rare community members, cells with irregular morphologies, small quantity samples, and studies that employ advanced optical microscopy.
Keywords: Contamination; Lab-on-a-chip; Laser; Microfluidic device; Multiple displacement amplification; Nanoliter; Optical tweezer; Single amplified genome; Single-cell sequencing; Whole-genome amplification.
© 2013 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

