Condensin aids sister chromatid decatenation by topoisomerase II
- PMID: 24062159
- PMCID: PMC3874195
- DOI: 10.1093/nar/gkt882
Condensin aids sister chromatid decatenation by topoisomerase II
Abstract
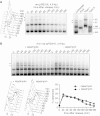
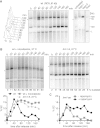
The condensin complex is a key determinant of mitotic chromosome architecture. In addition, condensin promotes resolution of sister chromatids during anaphase, a function that is conserved from prokaryotes to human. Anaphase bridges observed in cells lacking condensin are reminiscent of chromosome segregation failure after inactivation of topoisomerase II (topo II), the enzyme that removes catenanes persisting between sister chromatids following DNA replication. Circumstantial evidence has linked condensin to sister chromatid decatenation but, because of the difficulty of observing chromosome catenation, this link has remained indirect. Alternative models for how condensin facilitates chromosome resolution have been put forward. Here, we follow the catenation status of circular minichromosomes of three sizes during the Saccharomyeces cerevisiae cell cycle. Catenanes are produced during DNA replication and are for the most part swiftly resolved during and following S-phase, aided by sister chromatid separation. Complete resolution, however, requires the condensin complex, a dependency that becomes more pronounced with increasing chromosome size. Our results provide evidence that condensin prevents deleterious anaphase bridges during chromosome segregation by promoting sister chromatid decatenation.
Figures


References
-
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. - PubMed
-
- Strunnikov A, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. - PubMed
-
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

