Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules
- PMID: 24062337
- PMCID: PMC3787373
- DOI: 10.1083/jcb.201302139
Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules
Abstract
Temporal control of messenger RNA (mRNA) translation is an important mechanism for regulating cellular, neuronal, and developmental processes. However, mechanisms that coordinate timing of translational activation remain largely unresolved. Full-grown oocytes arrest meiosis at prophase I and deposit dormant mRNAs. Of these, translational control of cyclin B1 mRNA in response to maturation-inducing hormone is important for normal progression of oocyte maturation, through which oocytes acquire fertility. In this study, we found that dormant cyclin B1 mRNA forms granules in the cytoplasm of zebrafish and mouse oocytes. Real-time imaging of translation revealed that the granules disassemble at the time of translational activation during maturation. Formation of cyclin B1 RNA granules requires binding of the mRNA to Pumilio1 protein and depends on actin filaments. Disruption of cyclin B1 RNA granules accelerated the timing of their translational activation after induction of maturation, whereas stabilization hindered translational activation. Thus, our results suggest that RNA granule formation is critical for the regulation of timing of translational activation.
Figures
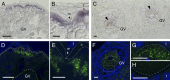
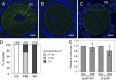


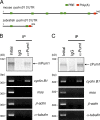
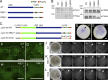


Comment in
-
Gene expression: RNA granules: the clock within.Nat Rev Mol Cell Biol. 2013 Nov;14(11):688. doi: 10.1038/nrm3684. Epub 2013 Oct 9. Nat Rev Mol Cell Biol. 2013. PMID: 24105321 No abstract available.
Similar articles
-
Pumilio1 phosphorylation precedes translational activation of its target mRNA in zebrafish oocytes.Zygote. 2018 Oct;26(5):372-380. doi: 10.1017/S0967199418000369. Epub 2018 Oct 5. Zygote. 2018. PMID: 30289101
-
Formation of mos RNA granules in the zebrafish oocyte that differ from cyclin B1 RNA granules in distribution, density and regulation.Eur J Cell Biol. 2016 Dec;95(12):563-573. doi: 10.1016/j.ejcb.2016.10.001. Epub 2016 Oct 4. Eur J Cell Biol. 2016. PMID: 27756483
-
A cis-acting element in the coding region of cyclin B1 mRNA couples subcellular localization to translational timing.Dev Biol. 2013 Oct 15;382(2):517-29. doi: 10.1016/j.ydbio.2013.05.014. Epub 2013 May 20. Dev Biol. 2013. PMID: 23701882
-
Post-transcriptional control of gene expression during mouse oogenesis.Results Probl Cell Differ. 2012;55:1-21. doi: 10.1007/978-3-642-30406-4_1. Results Probl Cell Differ. 2012. PMID: 22918798 Review.
-
Regulation of Translationally Repressed mRNAs in Zebrafish and Mouse Oocytes.Results Probl Cell Differ. 2017;63:297-324. doi: 10.1007/978-3-319-60855-6_13. Results Probl Cell Differ. 2017. PMID: 28779323 Review.
Cited by
-
Gene expression: RNA granules: the clock within.Nat Rev Mol Cell Biol. 2013 Nov;14(11):688. doi: 10.1038/nrm3684. Epub 2013 Oct 9. Nat Rev Mol Cell Biol. 2013. PMID: 24105321 No abstract available.
-
Synaptic control of local translation: the plot thickens with new characters.Cell Mol Life Sci. 2014 Jun;71(12):2219-39. doi: 10.1007/s00018-013-1506-y. Epub 2013 Nov 10. Cell Mol Life Sci. 2014. PMID: 24212248 Free PMC article. Review.
-
Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition.Cell Rep. 2014 Jun 12;7(5):1495-1508. doi: 10.1016/j.celrep.2014.05.002. Epub 2014 May 29. Cell Rep. 2014. PMID: 24882012 Free PMC article.
-
Cytoplasmic RNA Granules and Viral Infection.Annu Rev Virol. 2014 Nov;1(1):147-70. doi: 10.1146/annurev-virology-031413-085505. Annu Rev Virol. 2014. PMID: 26958719 Free PMC article.
-
The Xenopus Maternal-to-Zygotic Transition from the Perspective of the Germline.Curr Top Dev Biol. 2015;113:271-303. doi: 10.1016/bs.ctdb.2015.07.021. Epub 2015 Aug 21. Curr Top Dev Biol. 2015. PMID: 26358876 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

