The formin FHOD1 and the small GTPase Rac1 promote vaccinia virus actin-based motility
- PMID: 24062339
- PMCID: PMC3787377
- DOI: 10.1083/jcb.201303055
The formin FHOD1 and the small GTPase Rac1 promote vaccinia virus actin-based motility
Abstract
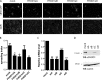
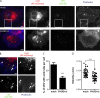
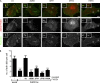
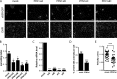
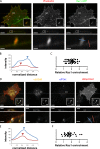
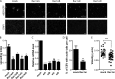
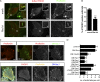
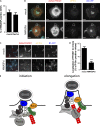
Vaccinia virus dissemination relies on the N-WASP-ARP2/3 pathway, which mediates actin tail formation underneath cell-associated extracellular viruses (CEVs). Here, we uncover a previously unappreciated role for the formin FHOD1 and the small GTPase Rac1 in vaccinia actin tail formation. FHOD1 depletion decreased the number of CEVs forming actin tails and impaired the elongation rate of the formed actin tails. Recruitment of FHOD1 to actin tails relied on its GTPase binding domain in addition to its FH2 domain. In agreement with previous studies showing that FHOD1 is activated by the small GTPase Rac1, Rac1 was enriched and activated at the membrane surrounding actin tails. Rac1 depletion or expression of dominant-negative Rac1 phenocopied the effects of FHOD1 depletion and impaired the recruitment of FHOD1 to actin tails. FHOD1 overexpression rescued the actin tail formation defects observed in cells overexpressing dominant-negative Rac1. Altogether, our results indicate that, to display robust actin-based motility, vaccinia virus integrates the activity of the N-WASP-ARP2/3 and Rac1-FHOD1 pathways.
Figures


Comment in
-
A role for the small GTPase Rac1 in vaccinia actin-based motility.Small GTPases. 2014;5(2):e29038. doi: 10.4161/sgtp.29038. Epub 2014 Oct 31. Small GTPases. 2014. PMID: 25483216 Free PMC article.
References
-
- Bernardini M.L., Mounier J., d’Hauteville H., Coquis-Rondon M., Sansonetti P.J. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA. 86:3867–3871 10.1073/pnas.86.10.3867 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

