Single-molecule FRET and linear dichroism studies of DNA breathing and helicase binding at replication fork junctions
- PMID: 24062430
- PMCID: PMC3808625
- DOI: 10.1073/pnas.1314862110
Single-molecule FRET and linear dichroism studies of DNA breathing and helicase binding at replication fork junctions
Abstract
DNA "breathing" is a thermally driven process in which base-paired DNA sequences transiently adopt local conformations that depart from their most stable structures. Polymerases and other proteins of genome expression require access to single-stranded DNA coding templates located in the double-stranded DNA "interior," and it is likely that fluctuations of the sugar-phosphate backbones of dsDNA that result in mechanistically useful local base pair opening reactions can be exploited by such DNA regulatory proteins. Such motions are difficult to observe in bulk measurements, both because they are infrequent and because they often occur on microsecond time scales that are not easy to access experimentally. We report single-molecule fluorescence experiments with polarized light, in which tens-of-microseconds rotational motions of internally labeled iCy3/iCy5 donor-acceptor Förster resonance energy transfer fluorophore pairs that have been rigidly inserted into the backbones of replication fork constructs are simultaneously detected using single-molecule Förster resonance energy transfer and single-molecule fluorescence-detected linear dichroism signals. Our results reveal significant local motions in the ∼100-μs range, a reasonable time scale for DNA breathing fluctuations of potential relevance for DNA-protein interactions. Moreover, we show that both the magnitudes and the relaxation times of these backbone breathing fluctuations are significantly perturbed by interactions of the fork construct with a nonprocessive, weakly binding bacteriophage T4-coded helicase hexamer initiation complex, suggesting that these motions may play a fundamental role in the initial binding, assembly, and function of the processive helicase-primase (primosome) component of the bacteriophage T4-coded DNA replication complex.
Keywords: DNA helicase; T4 primosome helicase; single-molecule linear dichroism; smFRET; thermal fluctuations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 is the mean square magnitude of the signal fluctuations,
is the mean square magnitude of the signal fluctuations,  is square average signal, and
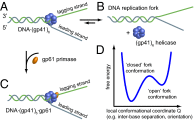
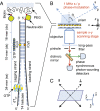
is square average signal, and  is the correlation time. The donor–acceptor chromophore labels are placed at the ss–ds fork junction of the DNA construct (
is the correlation time. The donor–acceptor chromophore labels are placed at the ss–ds fork junction of the DNA construct (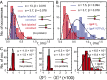
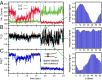
 ) obtained from the analysis of smFRET/smFLD trajectories for DNA fork constructs, which were labeled with iCy3/iCy5 placed deep in a duplex region (blue) or at the replication fork junction (red) in the absence of helicase protein. (B) A comparison is shown for a fork-labeled construct in the absence (red) and the presence (blue) of the “frozen” hexameric helicase (gp41 ∙ GTPγS)6 composed of 300 nM gp41 and 6 μM GTPγS. (C) A comparison is shown of histograms of the relative magnitudes
) obtained from the analysis of smFRET/smFLD trajectories for DNA fork constructs, which were labeled with iCy3/iCy5 placed deep in a duplex region (blue) or at the replication fork junction (red) in the absence of helicase protein. (B) A comparison is shown for a fork-labeled construct in the absence (red) and the presence (blue) of the “frozen” hexameric helicase (gp41 ∙ GTPγS)6 composed of 300 nM gp41 and 6 μM GTPγS. (C) A comparison is shown of histograms of the relative magnitudes  of the fluctuating smFLDr signal for duplex-labeled DNA (Left), fork-labeled DNA (Center), and fork-labeled DNA + (gp41 ∙ GTPγS)6 (Right). Histograms of the relaxation times were characterized using the gamma distribution function, given in the text, with skewedness and width parameters α (dimensionless) and β (in units of milliseconds), respectively.
of the fluctuating smFLDr signal for duplex-labeled DNA (Left), fork-labeled DNA (Center), and fork-labeled DNA + (gp41 ∙ GTPγS)6 (Right). Histograms of the relaxation times were characterized using the gamma distribution function, given in the text, with skewedness and width parameters α (dimensionless) and β (in units of milliseconds), respectively.Comment in
-
Watching DNA breath one molecule at a time.Proc Natl Acad Sci U S A. 2013 Oct 22;110(43):17173-4. doi: 10.1073/pnas.1316493110. Epub 2013 Oct 4. Proc Natl Acad Sci U S A. 2013. PMID: 24096577 Free PMC article. No abstract available.
References
-
- Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q Rev Biophys. 2002;35(4):431–478. - PubMed
-
- McConnell B, von Hippel PH. Hydrogen exchange as a probe of the dynamic structure of DNA. I. General acid-base catalysis. J Mol Biol. 1970;50(2):297–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

