FMRP regulates actin filament organization via the armadillo protein p0071
- PMID: 24062571
- PMCID: PMC3851716
- DOI: 10.1261/rna.037945.112
FMRP regulates actin filament organization via the armadillo protein p0071
Abstract
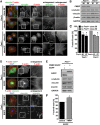
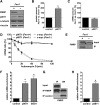
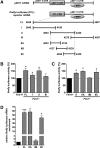
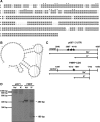
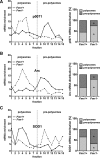
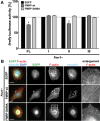
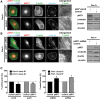
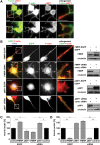
Loss of fragile X mental retardation protein (FMRP) causes synaptic dysfunction and intellectual disability. FMRP is an RNA-binding protein that controls the translation or turnover of a subset of mRNAs. Identifying these target transcripts is an important step toward understanding the pathology of the disease. Here, we show that FMRP regulates actin organization and neurite outgrowth via the armadillo protein p0071. In mouse embryonic fibroblasts (MEFs) lacking FMRP (Fmr1-), the actin cytoskeleton was markedly reorganized with reduced stress fibers and F-actin/G-actin ratios compared to fibroblasts re-expressing the protein. FMRP interfered with the translation of the p0071 mRNA in a 3'-UTR-dependent manner. Accordingly, FMRP-depleted cells revealed elevated levels of p0071 protein. The knockdown of p0071 in Fmr1- fibroblasts restored stress fibers and an elongated cell shape, thus rescuing the Fmr1- phenotype, whereas overexpression of p0071 in Fmr1+ cells mimicked the Fmr1- phenotype. Moreover, p0071 and FMRP regulated neurite outgrowth and branching in a diametrically opposed way in agreement with the negative regulation of p0071 by FMRP. These results identify p0071 as an important and novel FMRP target and strongly suggest that impaired actin cytoskeletal functions mediated by an excess of p0071 are key aspects underlying the fragile X syndrome.
Keywords: FMRP; actin organization; dendrite formation; neuromorphogenesis; p0071.
Figures
References
-
- Anastasiadis PZ, Reynolds AB 2001. Regulation of Rho GTPases by p120-catenin. Curr Opin Cell Biol 13: 604–610 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
