From simple to detailed models for cell polarization
- PMID: 24062577
- PMCID: PMC3785957
- DOI: 10.1098/rstb.2013.0003
From simple to detailed models for cell polarization
Abstract
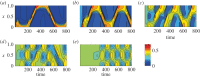
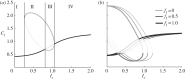
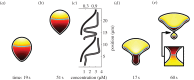
Many mathematical models have been proposed for the process of cell polarization. Some of these are 'functional models' that capture a class of dynamical behaviour, whereas others are derived from features of signalling molecules. Some mechanistic models are detailed, and therefore complex, whereas others are simplified. Each type contributes to our understanding of cell polarization. However, the huge variety at different levels of detail makes comparisons challenging. Here, we provide examples of both elementary and more detailed models for polarization. We also display how a recent mathematical method, local perturbation analysis, can provide an appropriate tool for such comparisons. This technique simplifies and speeds up the model development process by revealing the effect of model extensions, parameter variations and in silico manipulations such as knock-out or over-expression of key molecules. Finally, simulations in both one dimension and two dimensions, and particularly in deforming two-dimensional 'cells', can highlight behaviour not captured by traditional simulation methods.
Keywords: cell polarization; mathematical analysis; pattern formation; reaction–diffusion equations.
Figures
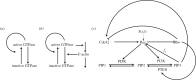
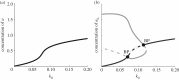
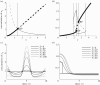
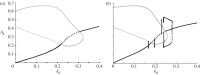

References
-
- Jilkine A, Edelstein-Keshet L. 2011. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput. Biol. 7, e1001121 (doi:10.1371/journal.pcbi.1001121) - DOI - PMC - PubMed
-
- Mori Y, Jilkine A, Edelstein-Keshet L. 2008. Wave-pinning and cell polarity from a bistable reaction–diffusion system. Biophys. J. 94, 3684–3697 (doi:10.1529/biophysj.107.120824) - DOI - PMC - PubMed
-
- Holmes W, Carlsson A, Edelstein-Keshet L. 2012. Regimes of wave type patterning driven by refractory actin feedback: transition from static polarization to dynamic wave behaviour. Phys. Biol. 9, 046005 (doi:10.1088/1478-3975/9/4/046005) - DOI - PMC - PubMed
-
- Holmes W, Lin B, Levchenko A, Edelstein-Keshet L. 2012. Modelling cell polarization driven by synthetic spatially graded Rac activation. PLoS Comput. Biol. 8, e1002366 (doi:10.1371/journal.pcbi.1002366) - DOI - PMC - PubMed
-
- Mareé AF, Jilkine A, Dawes A, Grieneisen VA, Edelstein-Keshet L. 2006. Polarization and movement of keratocytes: a multiscale modelling approach. Bull. Math. Biol. 68, 1169–1211 (doi:10.1007/s11538-006-9131-7) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

